Hard tissues like seashells and bone are made of proteins bound to minerals, but the process nature uses to create these materials is complex and not well understood. In this study, rather than directly untangle these byzantine protein–mineral interfaces, researchers used models. They selected a well-characterized mineral (mica) and then designed and synthesized proteins from scratch to systematically test how they assemble into a material. They brought their samples to the Advanced Light Source (ALS) to determine the finished structures.
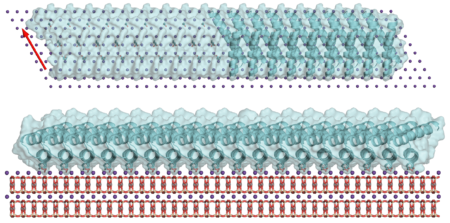
At Beamline 12.3.1, small-angle x-ray scattering (SAXS) revealed that the synthesized protein matched well with the natural protein in solution, taking a nanorod shape. The researchers designed the proteins to have repeating units that could slot into the mica lattice, with sites that could interact with the mica, other proteins, or both. The different-length proteins formed various structures on the mica surface, including a honeycomb-like pattern. When the researchers tested units that had a protein–mica interface but not a protein–protein interface, they expected the protein to bind only to the mica in simple formations. Surprisingly, the components formed extensive liquid-crystal arrays.
Computational protein design and structural determination via x-ray scattering are enabling scientists to characterize hard tissues to a greater extent than ever before. In addition, the ability to tune the final product by altering the design of the component proteins could be useful for devising new materials made of proteins and minerals.
H. Pyles, S. Zhang, J.J. De Yoreo, and D. Baker, “Controlling protein assembly on inorganic crystals through designed protein interfaces,” Nature 571, 251 (2019), doi:10.1038/s41586-019-1361-6.