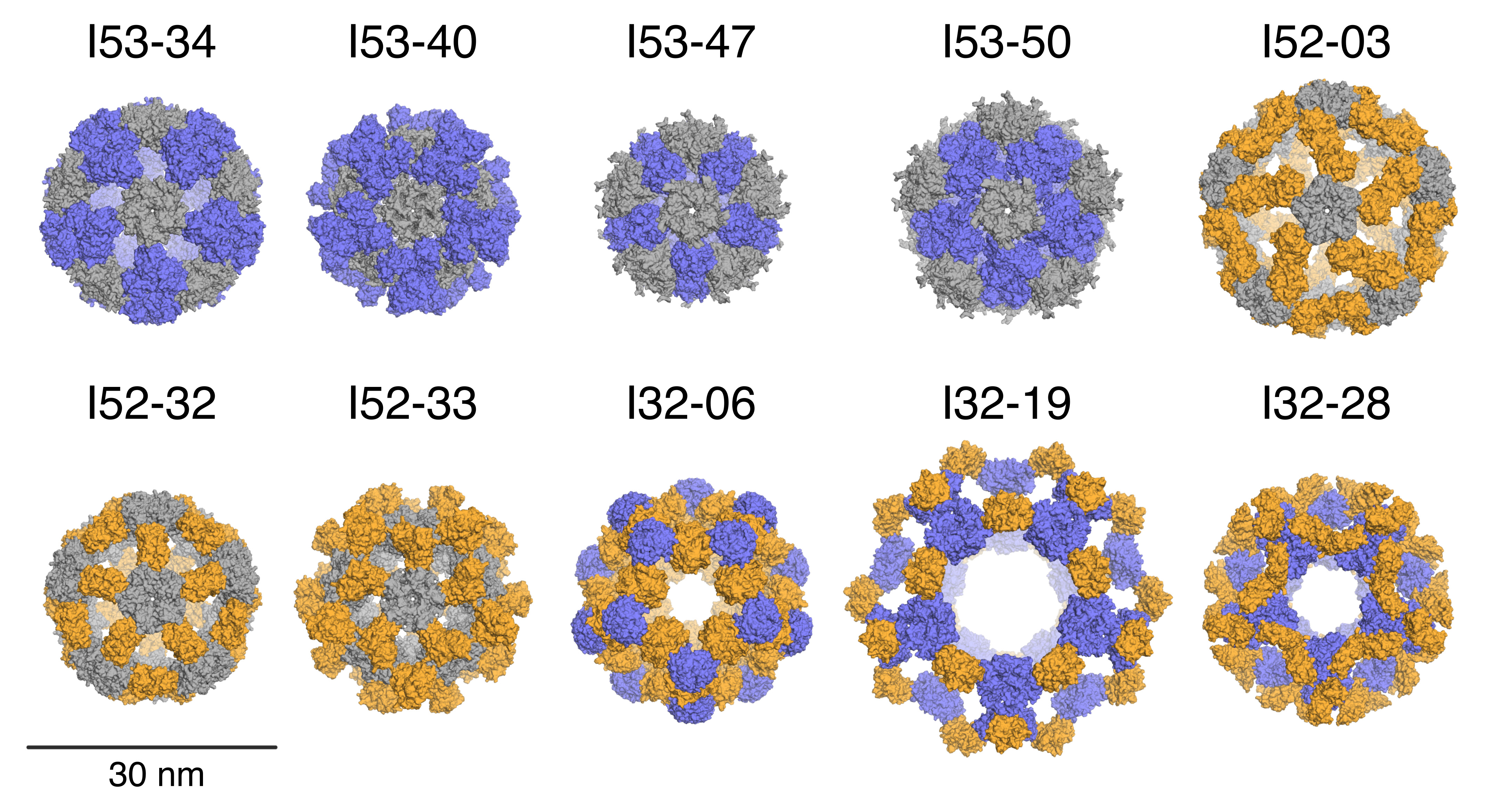
Nature provides many examples of self-assembling molecular machines, including icosahedral protein cages that serve as compartments for essential biochemical reactions and the protection and delivery of viral genomes. Now, researchers from the University of Washington have used computational methods to accurately design similar icosahedral protein complexes that self-assemble out of 120 modular pieces—60 each of two types of protein subunits. Hundreds of thousands of possible combinations of protein building blocks were systematically evaluated by computer as to their suitability for nanocage design. A few hundred of the nanocage designs were then produced by genetically reprogramming E. coli bacteria to express the desired proteins. Ultimately, ten designs survived the screening process to be characterized using electron microscopy, small-angle x-ray scattering (SAXS), and x-ray crystallography.
The mail-in SAXS program at ALS Beamline 12.3.1 is a relatively high-throughput characterzation method that doesn’t require protein crystallization. Specifically, the large, symmetric, nanocage complexes yielded feature-rich, distinct scattering profiles that closely matched the profiles calculated from the design models. The ability of the mail-in SAXS program to collect and analyze data quickly and reliably on many different samples makes it a powerful tool for the structure-validation pipeline.
The characteristics of the designed nanocages make them well suited for encapsulation of a broad range of materials, including small molecules, nucleic acids, polymers, and other proteins. These features, along with the precision and modularity with which they can be engineered, make these designed nanomaterials attractive starting points for new approaches to targeted drug delivery, vaccine design, and bioenergy.
SAXS performed at ALS Beamline 12.3.1.
J.B. Bale, S. Gonen, Y. Liu, W. Sheffler, D. Ellis, C. Thomas, D. Cascio, T.O. Yeates, T. Gonen, N.P. King, and D. Baker, “Accurate design of megadalton-scale two-component icosahedral protein complexes,” Science 353, 389 (2016).