While we humans view bacteria as the enemy, bacteria have enemies too, for example, viruses. To protect themselves, bacteria have developed an adaptive-type immune system that revolves around a unit of DNA known as CRISPR, which stands for Clustered Regularly Interspaced Short Palindromic Repeats. A CRISPR unit of DNA is made up of “repeat” elements, base-pair sequences ranging from 30 to 60 nucleotides in length, separated by “spacer” elements. The combination of CRISPR and CRISPR-associated (“Cas”) proteins enable bacteria to convert spacers into customized RNA molecules that silence critical portions of a foreign invader’s DNA. The CRISPR/Cas system also enables a bacterium to acquire immunity from similar invasions in the future by “remembering” prior infections based on the foreign DNA spacer elements integrated within the bacterium’s CRISPR loci.
Using macromolecular crystallography Beamline 8.3.1 at the ALS, a Berkeley research group studied x-ray crystal structures of CRISPR-associated Cas1 and Cas2 enzymes in Escherichia coli to see how foreign DNA is manipulated and bent upon being captured. Knowing how they function in bacterial genomes provides a possible mechanism for studying or correcting problems in human genomes.
They discovered that Cas1 and Cas2 function as molecular rulers that will not only recognize foreign DNA but also perfectly measure the DNA during the integration process. Cas1 and Cas2 capture double-stranded DNA whose ends are being separated by Cas1, which means that they could be possibly programed with DNA substrates containing a central double-stranded DNA region and single-stranded DNA on the ends. Afterwards, these DNA substrates could be inserted into specific sites along a target genome for editing purposes.
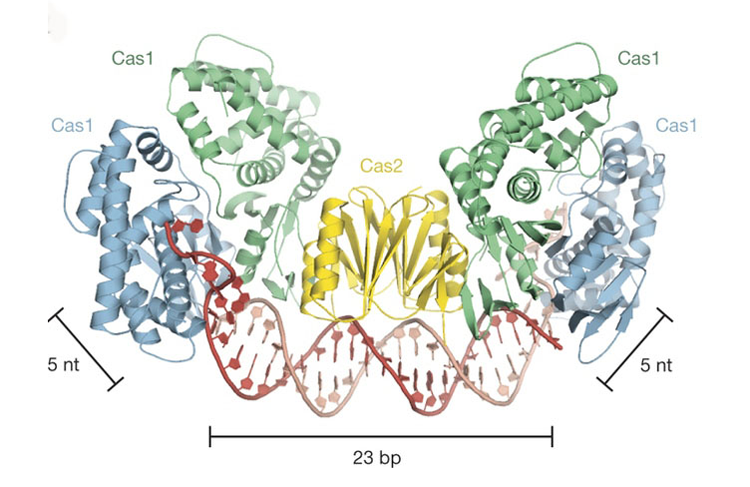
Work performed on ALS Beamline 8.3.1.
J.K. Nuñez, L.B. Harrington, P.J. Kranzusch, A.N. Engelman, and J.A. Doudna, “Foreign DNA capture during CRISPR–Cas adaptive immunity,” Nature 527, 535 (2015).