Large biological macromolecules (up to 10K atoms) can catalyze chemical reactions in ways that are difficult to replicate inorganically. Their size allows for the complexity needed to perform such functions, but it also makes them more susceptible to misfolding and aggregating. Thus, it’s important to understand the fundamental architectural principles that cause large proteins to favor specific conformations. At the nanoscale, where organic chemical groups interact with solvent water molecules, these principles are very different from the ones we use to build houses and cars (and yes, pyramids).
To investigate these principles, a group of researchers designed a protein that would self-assemble into a hollow pyramid (or tetrahedron). Upon crystallizing the macromolecule, the group found that they had indeed been successful in creating the assembly, but it was unexpectedly warped and collapsed in an asymmetric manner, with some edges bent inward—an asymmetric tetrahedron. To ensure that this was not an artifact of crystallization, they investigated the protein’s behavior in solution using small-angle x-ray scattering (SAXS) at ALS SIBYLS Beamline 12.3.1 (a joint crystallography and SAXS beamline). These and other studies showed that the collapse could be controlled by adjusting the salt concentration of the solution, and the structure could be disassembled by varying the pH. The flexibility of this macromolecule suggests that it could be useful for the controlled capture and release of smaller compounds.
Overall, the researchers expect that, with the tools and techniques developed here, the combination of SAXS with crystallography or electron microscopy could be increasingly useful in analyzing and optimizing designed protein assemblies and understanding their behavior in solution.
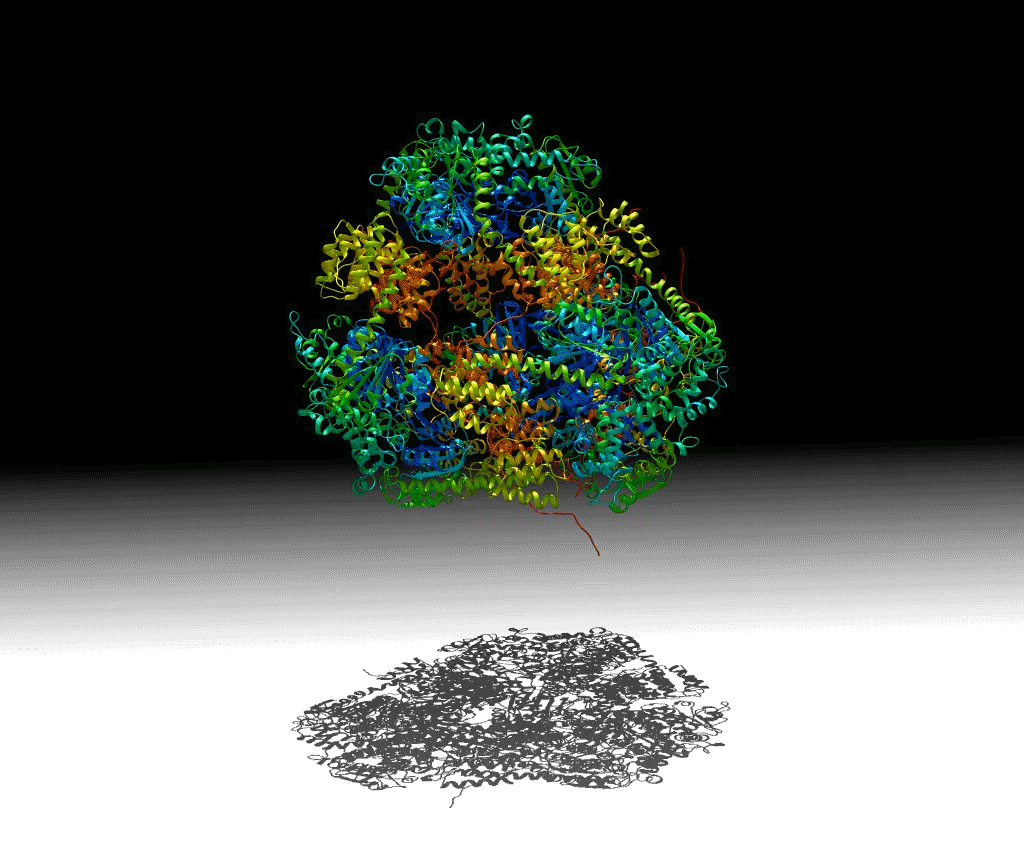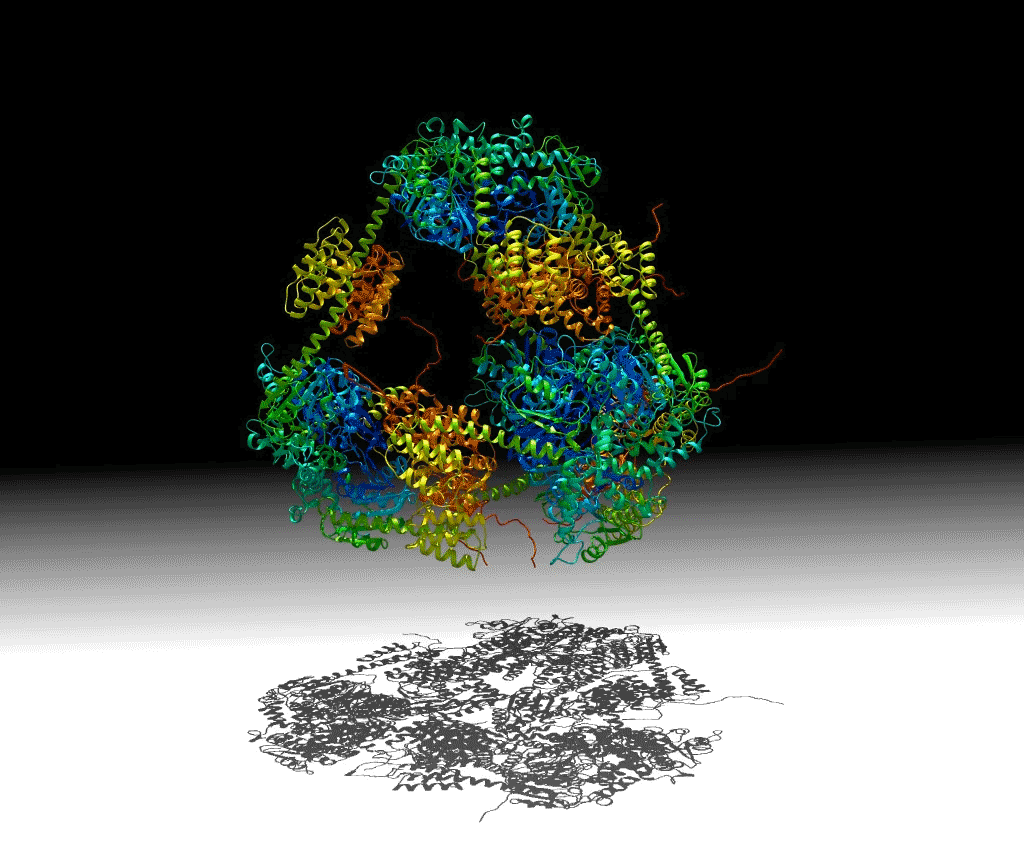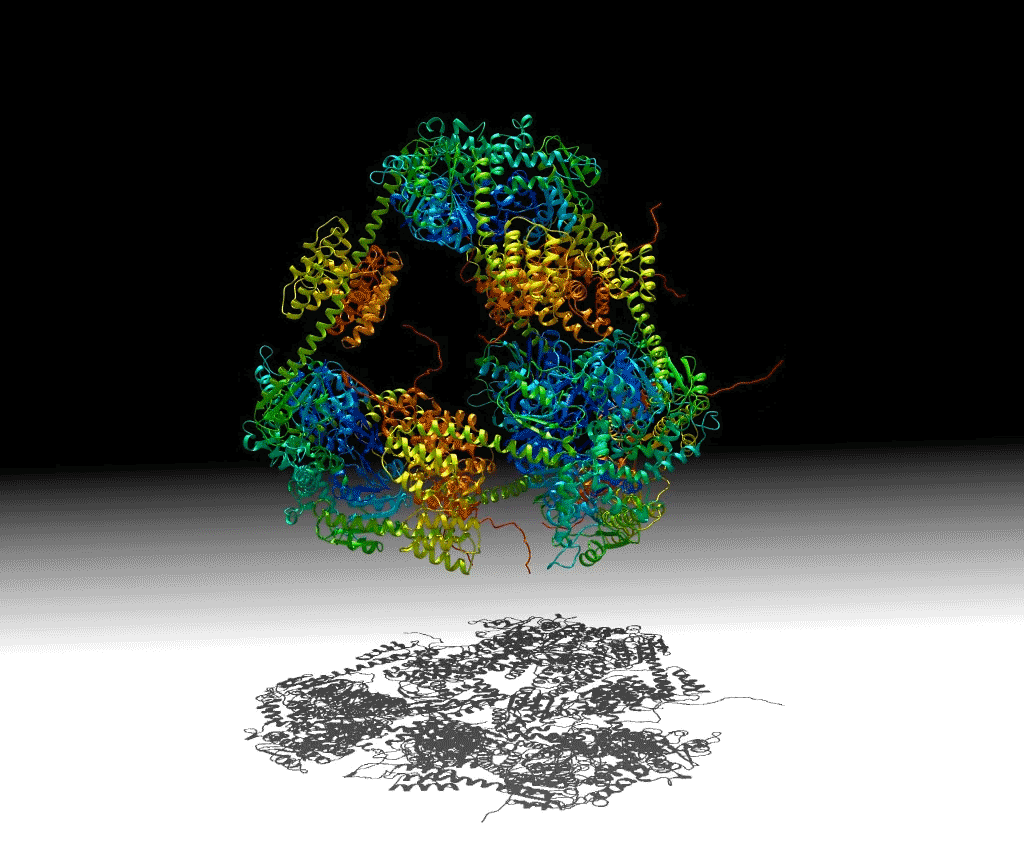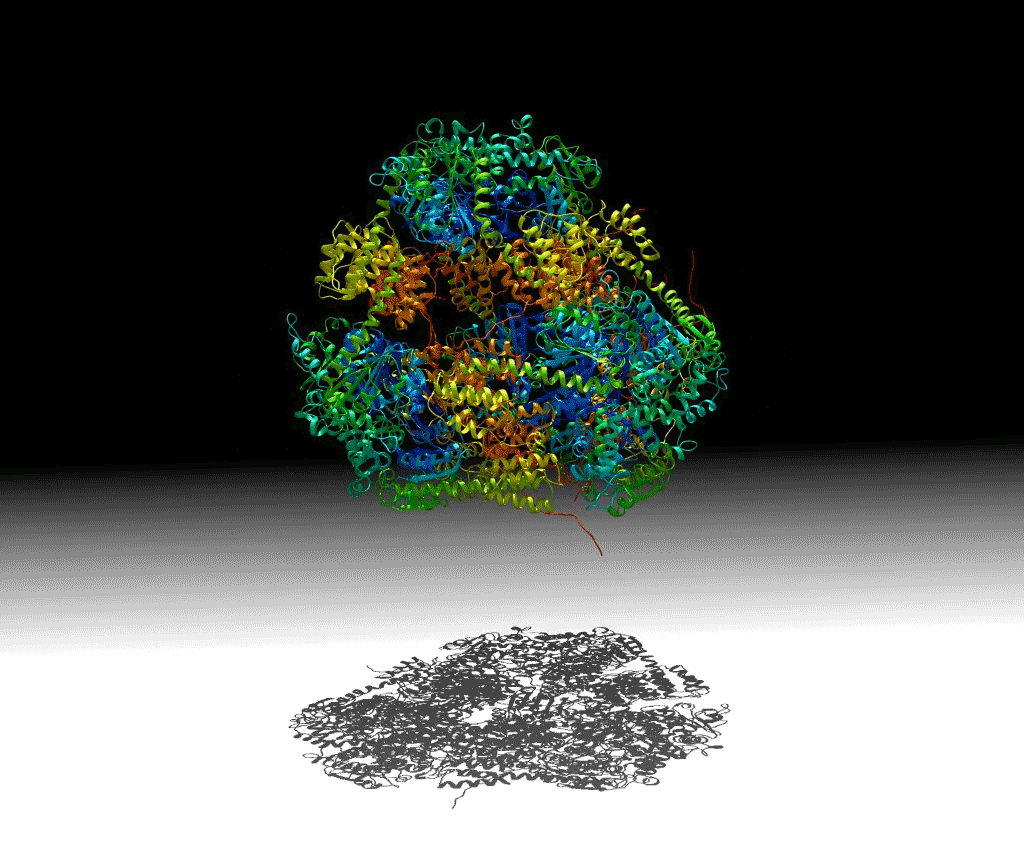
Work performed at ALS Beamline 12.3.1.
Y.-T. Lai, G.L. Hura, K.N. Dyer, H.Y.H. Tang, J.A. Tainer and T.O. Yeates, “Designing and defining dynamic protein cage nanoassemblies in solution,” Science Advances 2, e1501855 (2016).