SCIENTIFIC ACHIEVEMENT
Researchers successfully bioengineered changes to a molecular “assembly line” for bioactive compounds, based in part on insights gained from small-angle x-ray scattering at the Advanced Light Source (ALS).
SIGNIFICANCE AND IMPACT
The ability to re-engineer these assembly lines could improve their performance and facilitate the synthesis of new medically useful compounds.
Microbial peptide factories
Microbes are known to possess molecular “assembly lines” that produce an important class of compounds, many of which have uses as antibiotics, antifungals, and immunosuppressants. The compounds are peptides—chains of amino acids like RNA, but shorter and produced, not by ribosomes, but by cellular machines known as nonribosomal peptide synthetases (NRPSs).
Researchers have established that NRPSs are large, multi-enzyme clusters that synthesize compounds by passing a precursor molecule from one module to the next, with each “station” catalyzing an addition to the molecule. In the past decade, a great deal has been learned about how individual NRPS domains and modules work, but an understanding of how the assembly lines function as a whole has been lacking.
An atomic-scale view
In the hopes of eventually re-engineering NRPSs to make new and improved medicines, a team of researchers began investigating one, found in soil bacteria, that produces gramicidin, an antibiotic used to treat infected wounds, sore throats, and eye infections. Linear gramicidin synthetase is an NRPS with 16 modules that are physically attached to each other by flexible linkers or small docking domains. To gain insight into how individual modules relate to the whole, the researchers performed x-ray crystallography on linear gramicidin synthetase subunit A (LgrA), a two-module complex involved in peptide initiation and elongation. The resulting structures, obtained at the Canadian Light Source and the Advanced Photon Source, showed that the relative orientations of adjacent modules can vary dramatically, suggesting an unexpectedly high level of flexibility.
A structurally integrated view
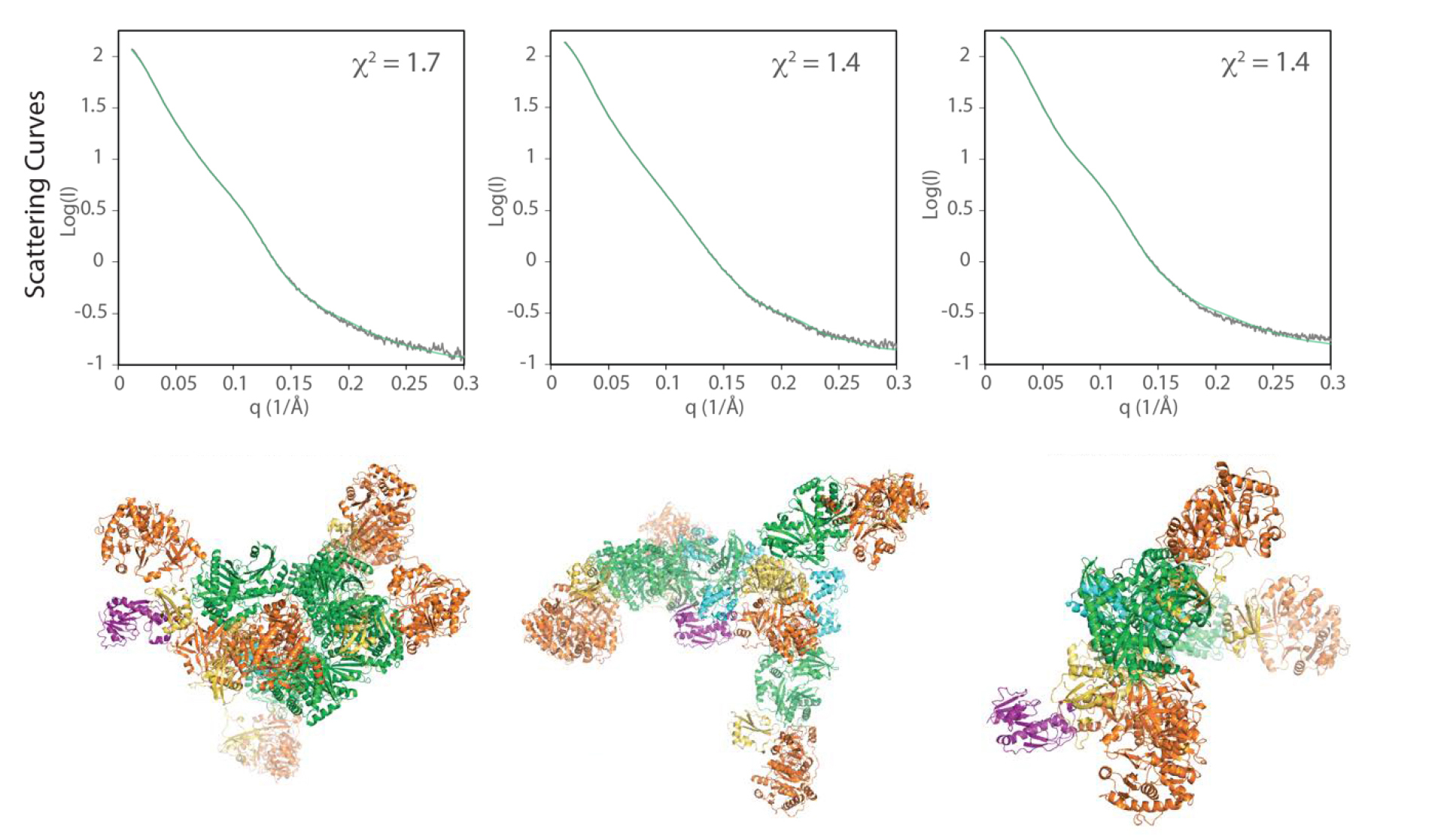
To assess whether the conformational variability seen in the crystal structures is an accurate reflection of what happens in a more natural state (i.e., in solution), the researchers analyzed the behavior of LgrA modules using small-angle x-ray scattering (SAXS) at ALS Beamline 12.3.1, the Structurally Integrated BiologY for Life Sciences (SIBYLS) beamline.
SAXS complements protein crystallography by enabling fast, accurate analysis of protein structures in solution at a resolution high enough to determine the protein’s shape, arrangement, and range of motion. Here, LgrA scattering patterns were used to select, from a random pool of thousands of independent structural models, an ensemble of models with theoretical scattering curves that fit well with the experimental data. The ensemble models were found to retain a level of structural flexibility similar to that of the original pool of independent models, consistent with the interpretation that LgrA is highly flexible. The results confirm that large conformational changes in LgrA as a whole are possible, even as individual modules perform their respective functions.
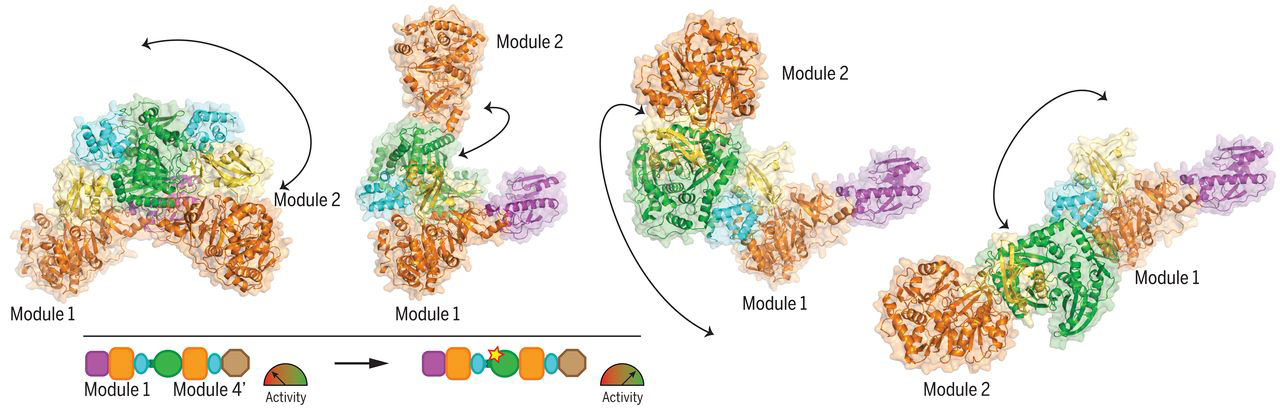
Chimeric LgrA with improved performance
Guided by insights derived from the structural data, together with analyses of the evolutionary interdependence of the domain interfaces, the researchers performed module-swapping experiments in which parts from a different NRPS were fused onto an LgrA module. Then, by introducing a couple of point mutations at the modular interface, they were able to double the activity of the module-swapped chimeric NRPS over the original. In the future, the researchers envision that this approach could be combined with other bioengineering strategies to help NRPSs fulfill their long-held promise as assembly lines for designer bioactive compounds.
Contact: Martin Schmeing
Researchers: J.M. Reimer, M. Eivaskhani, I. Harb, A. Guarné, and T.M. Schmeing (McGill University, Canada), and M. Weigt (Sorbonne University, France).
Funding: Canadian Institutes of Health Research, European Union Horizon 2020, Natural Sciences and Engineering Research Council of Canada, and Boehringer Ingelheim Fonds. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences Program.
Publication: J.M. Reimer, M. Eivaskhani, I. Harb, A. Guarné, M. Weigt, T.M. Schmeing, “Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility,” Science 366, 706 (2019), doi:10.1126/science.aaw4388.
Adapted from the Berkeley Lab Science Snapshot, “X-Ray Technology Sheds New Light on Antibiotic Synthesis.”
ALS SCIENCE HIGHLIGHT #416