SCIENTIFIC ACHIEVEMENT
Using x-ray crystallography at the Advanced Light Source (ALS), scientists characterized designed membrane proteins to better understand the forces that stabilize these large, complex structures.
SIGNIFICANCE AND IMPACT
The results necessitate a rethinking of membrane-protein biophysics and could lead to better therapies for related illnesses as well as functional membrane proteins for engineering applications.
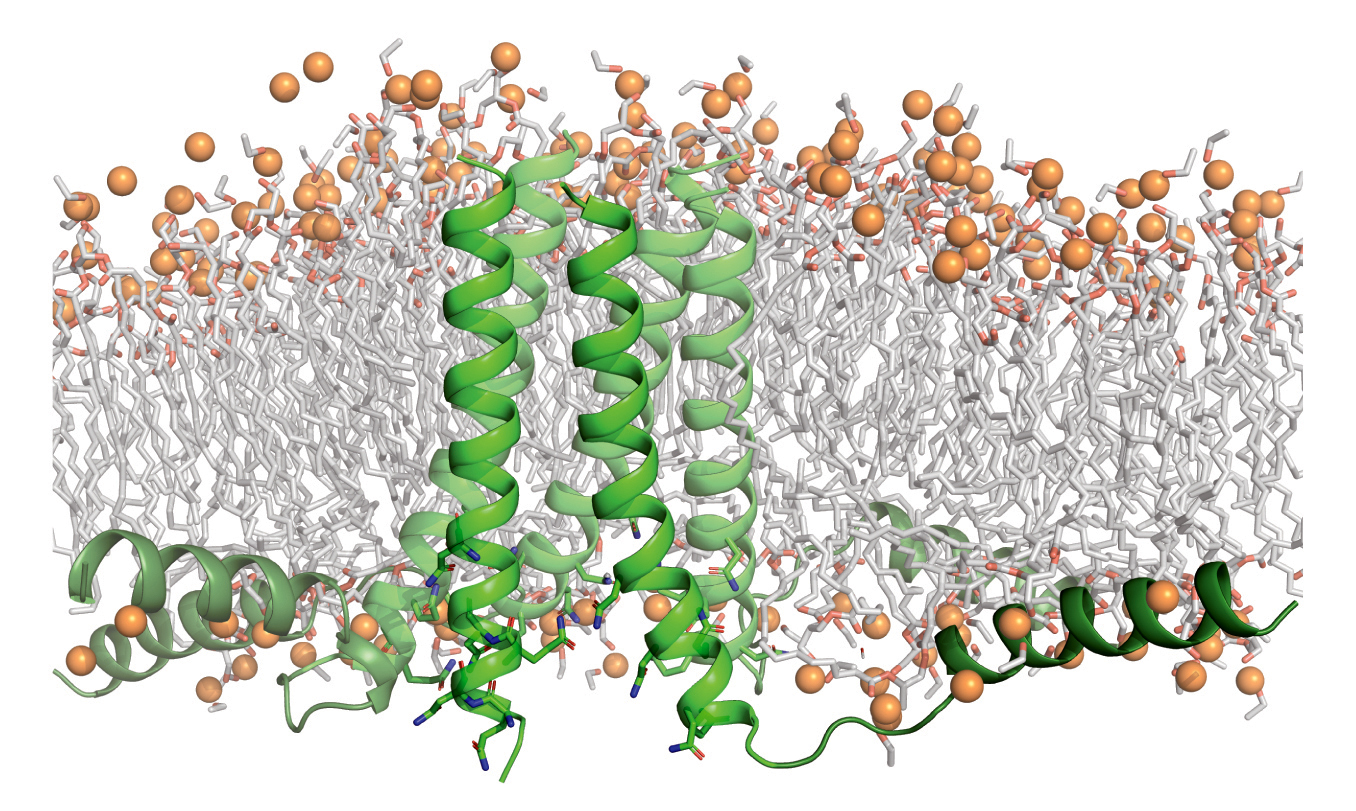
Biological gatekeepers
Certain proteins in cell membranes connect the cell’s complex inner workings with the dynamic outside world. Needed resources flow in, and wastes flow out. These membrane proteins, therefore, are essential for life. Genetic mutations that disturb the structural integrity and biological functions of membrane proteins cause many diseases (cancer, cystic fibrosis, and neurodegeneration, for example). Accordingly, over half of all drugs approved by the Food and Drug Administration target membrane-protein structures.
In light of this, researchers are interested in the general physics principles underlying how membrane proteins fold and stabilize their molecular structures. An atomic-level understanding would help in predicting and responding to mutations as well as in engineering synthetic membrane proteins with new useful functions.
Role of van der Waals forces
A long-standing debate among scientists is whether van der Waals forces—short-range forces between uncharged molecules—are too weak to stabilize large, complex, three-dimensional (3D) membrane proteins. Such forces are critical to maintaining the structural integrity of water-soluble proteins floating freely inside a cell. But within a cell membrane (consisting of oily lipids and very little water), it’s unclear whether van der Waals forces play an active role or whether they take a back seat to hydrogen bonds, salt bridges, and spatial constraints. Many synthetic membrane proteins have been designed using polar interactions, metal-ligand interactions, and other strategies to induce folding. Yet none have been successfully designed (and visualized at high resolution) based primarily on van der Waals interactions.
Variations on a natural protein
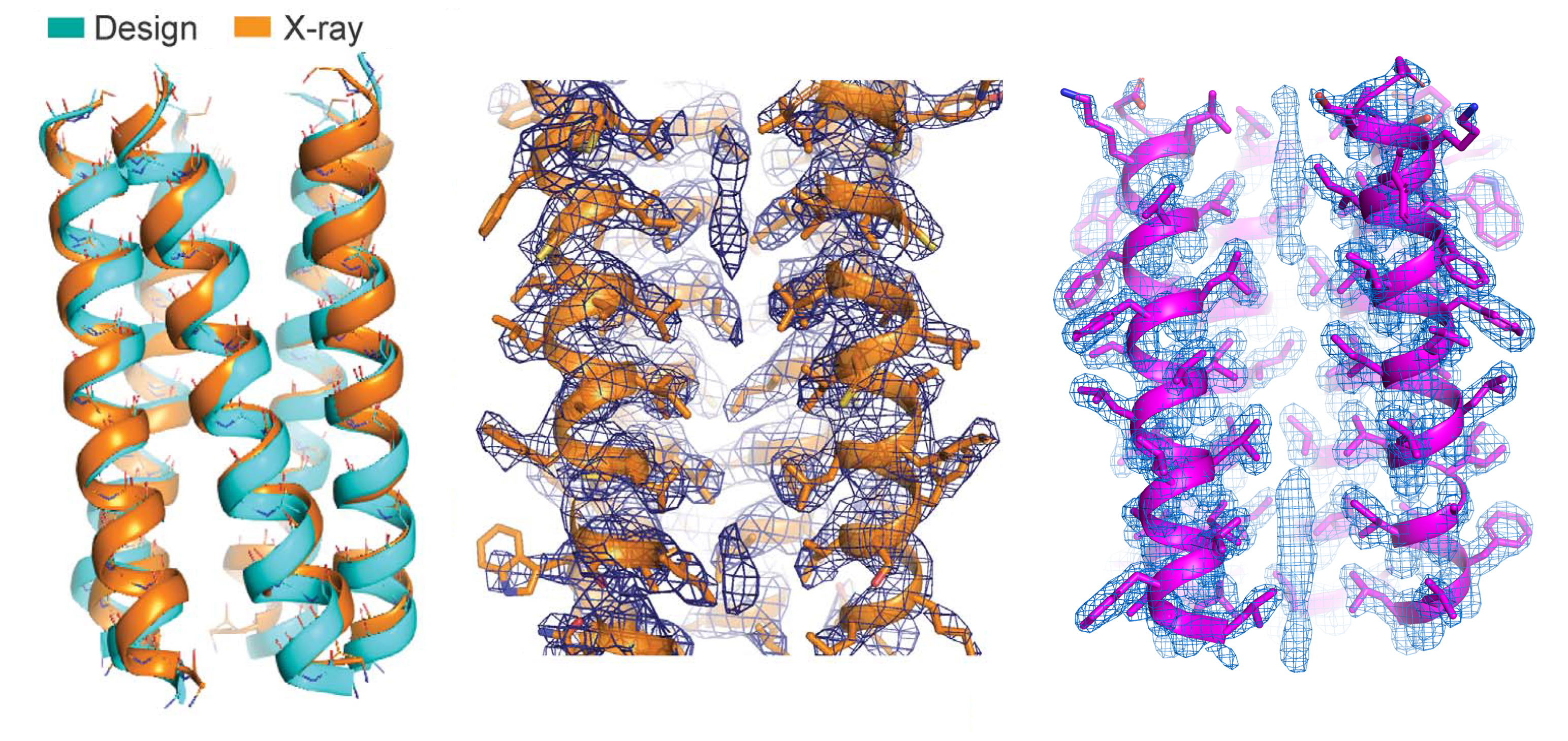
Here, researchers synthesized proteins based on a natural protein, phospholamban (PLN), an important regulator of calcium flux in the heart. Using computer models and molecular dynamics simulations, a variant of the PLN transmembrane domain was designed with two key mutations that stabilized its fold and 3D structures with extensive van der Waals interactions and side-chain steric packing (i.e., amino-acid puzzle-piecing). To verify the synthesized protein’s structure, the researchers used high-resolution protein crystallography at ALS Beamline 8.3.1.
The results showed that the PLN variant closely matched the computer model and that its structural stability was indeed due to side-chain steric packing and van der Waals interactions. Based on this structure, the researchers defined a basic pattern of side-chain packing that optimizes attractive van der Waals interactions favorable to folding.
A stringent packing code
Using the pattern as a guideline, the researchers then designed fully synthetic membrane proteins, verifying the structure of one of them at Beamline 8.3.1. Biochemical tests showed that the proteins were extremely stable, under conditions that would damage most proteins. However, even small deviations from the prescribed packing code caused the proteins to dismantle. Thus, van der Waals forces can serve as the sole source of structural integrity in membrane proteins—even leading to hyperstability—but the line between hyperstability and complete instability is razor thin. This is vastly different from how researchers previously perceived these forces in membrane proteins and from how hydrophobic steric packing is known to operate in water-soluble proteins. The results should be useful in predicting membrane-protein structures and the effects of mutations, as well as in the design of more complex and functional membrane proteins for engineering applications.
Contact: Marco Mravic
Researchers: M. Mravic, J.L. Thomaston, M. Tucker, P.E. Solomon, and W.F. DeGrado (University of California, San Francisco) and L. Liu (Peking University Shenzhen and DLX Scientific, USA).
Funding: National Institutes of Health, National Science Foundation, Howard Hughes Medical Institute, and University of California Office of the President. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences Program (DOE BES).
Publication: M. Mravic, J.L. Thomaston, M. Tucker, P.E. Solomon, L. Liu, and W.F. DeGrado, “Packing of apolar side chains enables accurate design of highly stable membrane proteins,” Science 363, 1418 (2019), doi:10.1126/science.aav7541.
ALS SCIENCE HIGHLIGHT #397