SCIENTIFIC ACHIEVEMENT
Researchers discovered two antibodies that can prevent the protein interactions responsible for severe malaria and elucidated their binding mechanism at the Advanced Light Source (ALS).
SIGNIFICANCE AND IMPACT
These broadly reactive antibodies are likely to represent a common mechanism of acquired immunity to severe malaria and offer novel insights for vaccine design or targeted treatment.
Outsmarting malaria’s molecular disguise
Malaria causes approximately 600,000 deaths each year, mainly among young children living in sub-Saharan Africa, and ten times as many suffer from severe forms of the disease that result in long-lasting health and socioeconomic consequences. In the last few years, malaria progress has largely plateaued and emerging vaccines lack sufficient success rates, necessitating new interventions to combat this disease.
An important interaction that underlies severe malaria is the binding between the endothelial protein C receptor (EPCR) to CIDRα1, a specific domain within a key protein expressed on the surface of infected red blood cells. However, CIDRα1 has evolved extensive amino acid sequence diversity, enabling it to appear in thousands of variants and hampering vaccine development that targets its interaction with EPCR. In this study, researchers identified two antibodies that can inhibit the interaction between EPCR and CIDRα1 despite this sequence diversity and structurally characterized their mechanism for preventing this interaction.
The receptor interceptor
The researchers isolated B cells—white blood cells that produce antibodies to fight infections—from three Ugandan adults with acquired immunity to malaria. B cells that reacted to either of the two most diverse variants of CIDRα1 were then cultured to produce antibodies, and these antibodies were further screened to identify their ability to interrupt the interaction between EPCR and different variants of CIDRα1. The monoclonal antibodies C7 and C74 were found to be the most effective in blocking the binding of CIDRα1 and EPCR, preventing the interaction across a wide range of CIDRα1 variants.
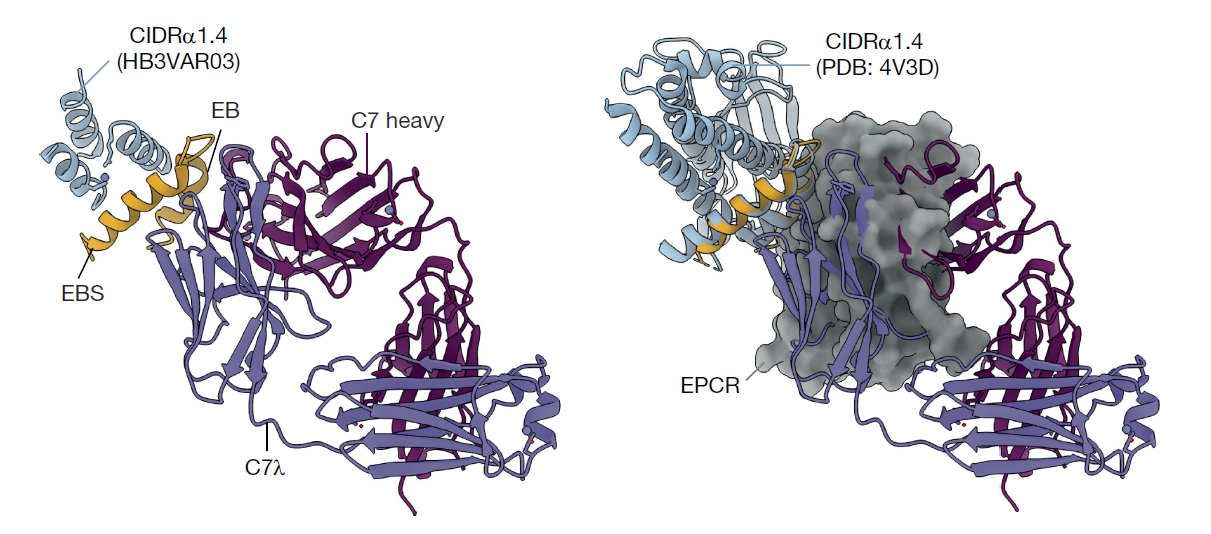
Using x-ray crystallography at ALS Beamline 5.0.2, the researchers characterized C7 in complex with CIDRα1.4, a variant of CIDRα1. Despite the small crystal used for this experiment, the high flux of the ALS enabled detailed structural analysis with high resolution. By overlaying the structural model of C7 in complex with CIDRα1.4 with that of EPCR in complex with the same CIDRα1 variant, the researchers found that C7 directly blocks binding to EPCR.
When researchers overlaid the ALS data with electron microscopy structural data, they found that C7 interacts with two different variants of CIDRα1 in nearly identical manners. The antibody targets conserved sites—regions that are similar across different variants—which explains how it can broadly prevent the interaction between CIDRα1 and EPCR despite the wide amino acid sequence diversity in CIDRα1.
Towards a new vaccine
Although C74 was derived from a different individual than C7 and has a different structure, it was found that the sites bound by both antibodies were very similar. There are only three amino acid residues that are conserved across different variants of CIDRα1, yet these are the key sites used by both C7 and C74 to bind to CIDRα1. Incredibly, the immune response of two different individuals developed the same strategy to prevent the interaction between CIDRα1 and EPCR.
Now that the researchers understand the minimal epitope, or minimal region, that these two broadly reactive antibodies recognize in CIDRα1, the next step is to design small antigens that mimic this surface area and direct the immune response to the conserved features of this epitope. These can then be used as immunogens to elicit antibodies similar to C7 and C74, with the goal of developing a malaria vaccine that protects against the most severe complications of this disease.
Contact: Evelien Bunnik
Researchers: R.A. Reyes, S. Bol, E. Martinez-Scholze, and E.M. Bunnik (University of Texas Health Science Center at San Antonio); S.S.R. Raghavan (Scripps Research Institute and University of Copenhagen and Righospitalet, Denmark); N.K. Hurlburt and M. Pancera (Fred Hutchinson Cancer Center); V. Introini, M. Gestal-Mato, B. López-Gutiérrez, S. Sanz, C. Bancells, and M. Bernabeu (European Molecular Biology Laboratory Barcelona); I.H. Kana, R.W. Jensen, T.G. Theander, L.Turner, and T. Lavstsen (University of Copenhagen and Righospitalet, Denmark); M.L. Fernández-Quintero, J.R. Loeffler, J.A. Ferguson, W.-H. Lee, G.M. Martin, and A.B. Ward (Scripps Research Institute); J.P. A. Lusingu and D.T.R. Minja (Tanga Research Centre, Tanzania); I. Ssewanyana (Infectious Disease Research Collaboration, Uganda); and M.E. Feeney and B. Greenhouse (University of California San Francisco).
Funding: National Institutes of Health, Howard Hughes Medical Institute, Lundbeck Foundation, Independent Research Fund Denmark, Kirsten & Freddy Johansens Fond, Novo Nordisk Fonden, Marie Skłodowska-Curie Actions fellowship, European Molecular Biology Laboratory, University of Texas Health Science Center at San Antonio, and Cancer Prevention and Research Institute of Texas. Operation of the ALS is supported by the US Department of Energy, Office of Science, Basic Energy Sciences program.
Publication: R.A. Reyes, S.S.R. Raghavan, N.K. Hurlburt, V. Introini, S. Bol, I.H. Kana, R.W. Jensen, E. Martinez-Scholze, M. Gestal-Mato, B. López-Gutiérrez, S. Sanz, C. Bancells, M.L. Fernández-Quintero, J.R. Loeffler, J.A. Ferguson, W.-H. Lee, G.M. Martin, T.G. Theander, J.P.A. Lusingu, D.T.R. Minja, I. Ssewanyana, M.E. Feeney, B. Greenhouse, A.B. Ward, M. Bernabeu, M. Pancera, L.Turner, E.M. Bunnik, and T. Lavstsen, “Broadly inhibitory antibodies to severe malaria virulence proteins,” Nature 636, 182 (2024), doi:10.1038/s41586-024-08220-3.
ALS SCIENCE HIGHLIGHT #516