Researchers have used a powerful soft x-ray microscope at the ALS to capture tomographic images of the genetic material in the nuclei of nerve cells at different stages of maturity. This study, the first quantitative analysis of nuclear organization in intact mammalian cells, has generated detailed 3D visualizations that show an unexpected connectivity in the genetic material, called chromatin. The results could help us understand how the patterning and reorganization of chromatin relate to the specialization of a stem cell’s function as specific genes are activated or silenced.
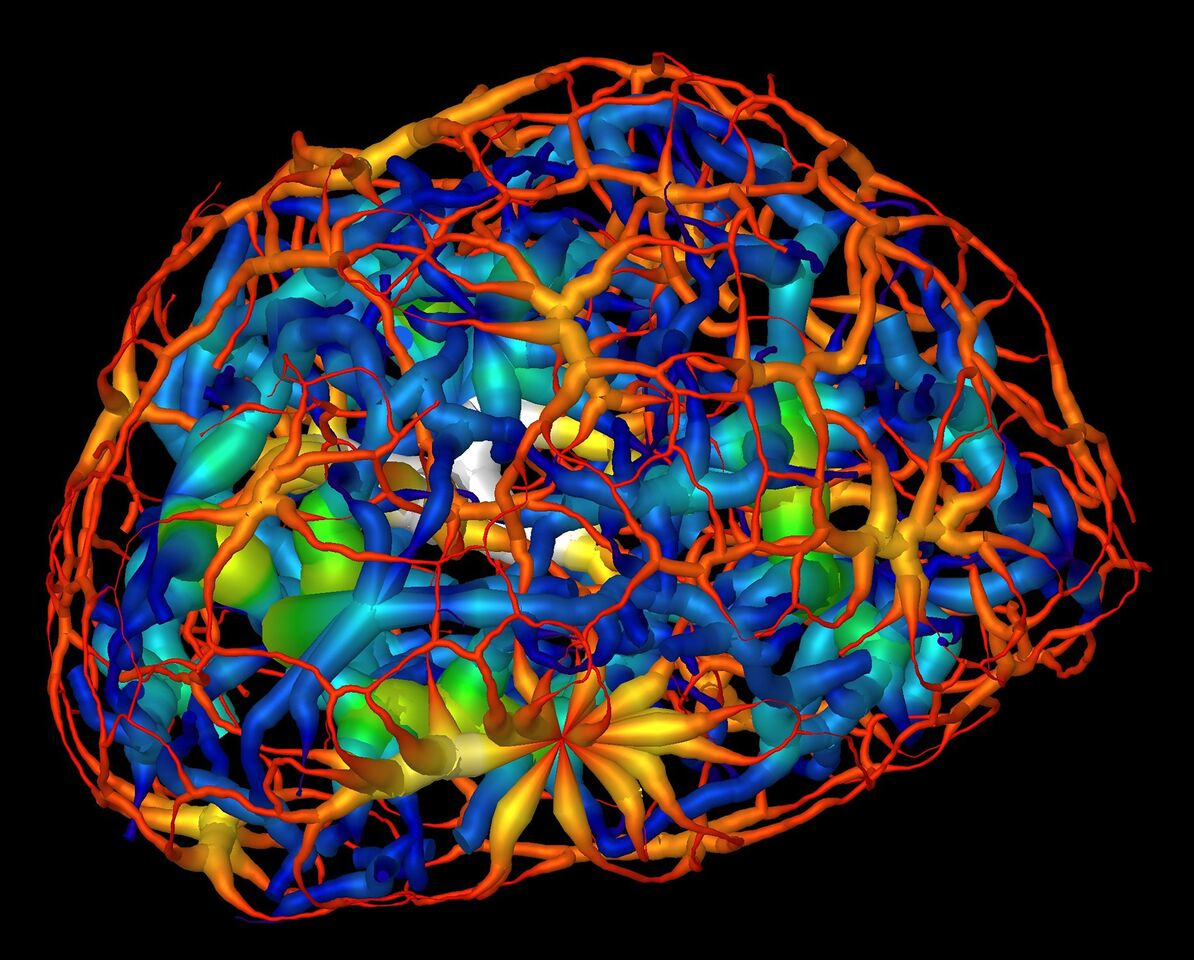
During cell division, chromatin, which consists of DNA, RNA, and other proteins, is compacted to form the chromosomes that pass along an organism’s genetic fingerprint to newly formed cells. The distribution of chromatin in the nucleus differs between cell types and developmental stages, suggesting that nuclear organization serves regulatory functions. However, understanding the logic of nuclear architecture and how it contributes to stem-cell differentiation remains challenging. Until now, it has only been possible to image the nucleus indirectly, by staining it, in which case the researcher has to take a leap of faith that the stain was evenly distributed. Chromatin is notoriously sensitive to chemical stains and other chemical additives that are often used in biological imaging to highlight regions of interest in a given sample.
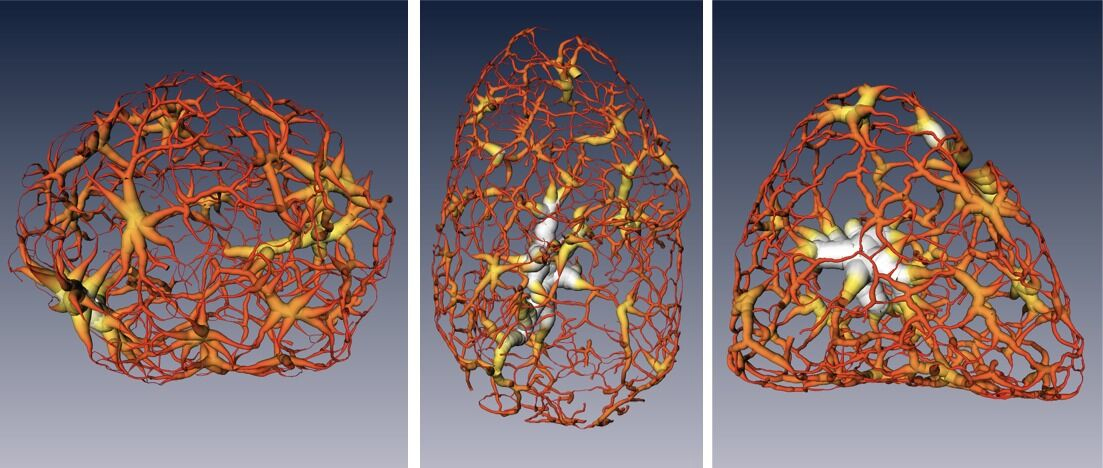
In this work, researchers used soft x-ray microtomography at ALS Beamline 2.1 (part of the National Center for X-Ray Tomography) to record a series of images from mouse olfactory nerve cells in three separate stages of development. The technique, which is unique to the ALS, provides a new way of looking at the nucleus without the need to chemically treat the cell, allowing visualization of intact cells in a near-native state at a resolution of about 50 nanometers.
Frozen cells at each stage were imaged from dozens of different angles, and each set of 2D images was used to calculate a 3D reconstruction detailing the changing chromatin formations in the nucleus. The images were collected using soft x-rays within the “water window” (284–543 eV), where biomolecules absorb x-rays an order of magnitude more than the surrounding water. The absorption is linear with biochemical composition and concentration, generating a unique x-ray linear absorption coefficient measurement for each voxel (3D pixel analog). Thus, the researchers were able to visually distinguish between different types of chromatin: heterochromatin, due to an increased biomolecular concentration, appears darker than euchromatin in computer-generated tomographic orthoslices (virtual sections) through the nucleus.
The results showed that chromatin compaction increases as the cell matures, and that condensed chromatin moves to the nuclear core during differentiation. Surprisingly, while it was previously thought that the chromatin existed as a series of disconnected islands, the results showed how the chromatin was compartmentalized into two distinct regions of “crowding” that form a continuous network throughout the nucleus. Also, based on comparison of these results to those of similar cells in which the gene for HP1β (a heterochromatin binding protein) had been inactivated (“knocked out”), the researchers concluded that HP1β regulates the reorganization of chromatin in mature neurons.
Soft x-ray tomography provides a powerful method to study chromatin and nuclear architecture in vivo. There is no need for chemical fixation and sectioning, preventing a plethora of artifacts introduced either by fixatives or by visualization of only thin sections. With the proven success of the imaging technique, the researchers believe it is possible to perform statistical analyses based on large collections of cell nuclei images sorted by different stages of development. Coupled with other types of imaging techniques, researchers hope to isolate individual gene-selection processes in upcoming work.
Contact: Carolyn Larabell
Research conducted by: M.A. Le Gros and C.A. Larabell (UC San Francisco and Berkeley Lab); E.J. Clowney, E. Markenscoff-Papadimitriou, B. Colquitt, and S. Lomvardas (UC San Francisco); A. Magklara (Foundation for Research and Technology-Hellas, Greece); A. Yen and M. Kellis (Broad Institute and MIT); and M. Myllys (Univ. of Jyväskylä, Finland).
Research funding: National Institutes of Health and the U.S. Department of Energy (DOE), Office of Biological and Environmental Research. Operation of the ALS is supported by the DOE Office of Basic Energy Sciences.
Publication about this research: M.A. Le Gros, E.J. Clowney, A. Magklara, A. Yen, E. Markenscoff-Papadimitriou, B. Colquitt, M. Myllys, M. Kellis, S. Lomvardas, and C.A. Larabell, “Soft x-ray tomography reveals gradual chromatin compaction and reorganization during neurogenesis in vivo,” Cell Reports 17, 2125 (2016). dx.doi.org/10.1016/j.celrep.2016.10.060
SCIENCE HIGHLIGHT #346