SCIENTIFIC ACHIEVEMENT
The Advanced Light Source (ALS) characterized a protein from a modern shark gene that explains the evolution of the adaptive immune system shared by all vertebrates.
SIGNIFICANCE AND IMPACT
Understanding the emergence of the adaptive immune system may aid researchers in advancing immunology, genetics, and biotechnology.
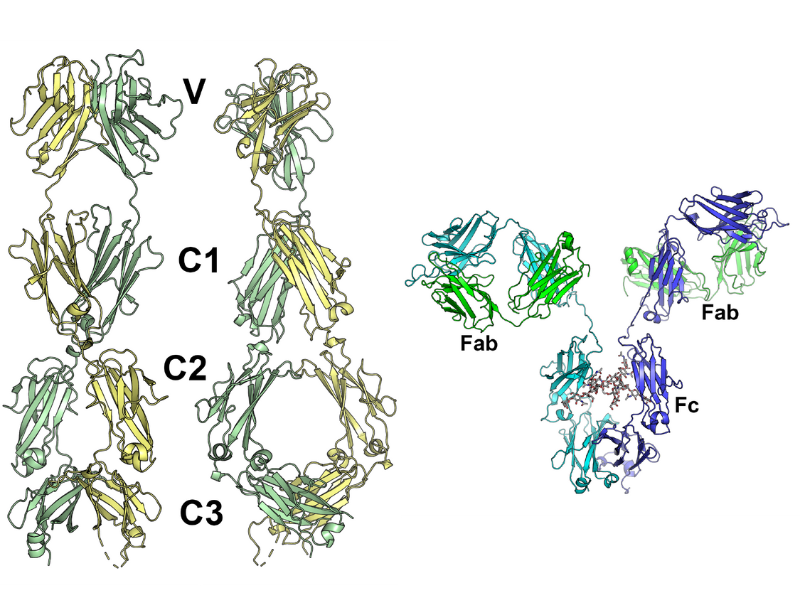
Left: The crystallographic model of the N-terminus of the UrIg2 protein from a nurse shark. Right: An example of one modern human antibody (IgG) whose variable region gene undergoes rearrangement.
The rise of adaptive immunity
Humans defend against infections through both the innate and adaptive immune systems. The innate response provides the first line of rapid defense, but it lacks both a way to address specific pathogens and a memory response to launch against attack by a returning invader. The adaptive immune system acts as a second line of defense. It lags behind the innate system because it must construct the antibodies to fight specific pathogens. The strength of this dual approach lies in the memory retained in the cells that produce antibodies that can be recalled to neutralize a returning threat.
The adaptive immune system is shared by all vertebrates and is believed to have developed soon after a genome-wide duplication event that occurred approximately 500 million years ago. The scientific community theorizes that the adaptive immune system developed when a mobile genetic element from a microbe—a recombination-activating gene (RAG) transposon—inserted itself into and split a gene in a eukaryotic cell, likely a white blood cell.
This random event led to a monumental and life-altering outcome. The process brought the repetitive elements from the transposon into this fractured gene with the RAG enzymes, which sparked the generation of an incalculable number of new proteins. To repair the fracture, the cell called in specialized machinery—double strand break repair enzymes—to fix the broken strands of genetic material. Proteins encoded by such “rearranged” genes eventually became antibodies—the front line of defense in the adaptive immune response.
Shedding light on a primordial gene
The ancient gene that set this unique course of events in motion was seemingly lost to time. That is until, a team of researchers working on the genomes of several shark species found a non-rearranging antibody-like gene (UrIg) in the major histocompatibility complex (MHC), a set of genes that encode the surface proteins that bind to pathogens in preparation for a response by specialized white blood cells (T lymphocytes) involved in the immune response.
In this new study, an ancient nonrearranging paralogue of UrIg, UrIg2, was also found in sharks. Incredibly, UrIg2 was shown to be encoded adjacent to a group of rearranging antibody and T cell receptor genes and is the best candidate to have been invaded by the RAG transposon.
The team at Scripps Research Institute expressed, purified, and crystallized the four N-terminal domains of the nurse shark UrIg2 protein. Using x-ray protein crystallography at Beamline 5.0.2 at the ALS, the researchers determined a three-dimensional structural model of the protein. The high flux of the ALS enabled high-resolution diffraction from the crystal that allowed for a detailed structural analysis.
With a structure resolved, the team had the missing piece of a puzzle that solidifies the genetic and phylogenetic story of immunity. The structure showed definitively that UrIg2 is a member of the antigen receptor family—each of the first four immunoglobulin superfamily domains most related to antibodies and/or T cell receptors.
Evolutionary perspective, advancing future studies
The researchers identified the candidate gene that catalyzed the initiation of the adaptive immune system. Further, the research team proposed a unified model for the evolution of adaptive immunity that is shared by all vertebrates.
This discovery provides valuable insights into the origins and mechanisms of how the adaptive immune system developed, deepening the scientific community’s understanding of the genetic basis of immune function that may have potential applications in medicine and biotechnology.
Contacts: Martin Flajnik and Robyn Stanfield
Researchers: M.F. Flajnik and Y. Ohta (University of Maryland, Baltimore), R.L. Stanfield and I.A. Wilson (Scripps Research Institute, La Jolla), A. Verissimo and A. Muñoz-Mérida (University of Porto), H.R. Neely (Harvard Medical School), and M.F. Criscitiello (Texas A&M University, College Station)
Funding: National Institutes of Health, the Norte Portugal Regional Operational Programme, and Fundação para a Ciência e Tecnologia (Portugal). Operation of the Berkeley Center for Structural Biology is supported in part by the Howard Hughes Medical Institute. Operation of the ALS is supported by the US Department of Energy, Office of Science, Basic Energy Sciences program.
Publication: M.F. Flajnik, R.L. Stanfield, A. Verissimo, H.R. Neely, A. Muñoz-Mérida, M.F. Criscitiello, I.A. Wilson, and Y. Ohta, “Origin of immunoglobulins and T cell receptors: A candidate gene for invasion by the RAG transposon,” Science Advances 11, eadw1273 (2025). doi: 10.1126/sciadv.adw1273
ALS SCIENCE HIGHLIGHT #528