Proteins consisting of identical subunits arranged symmetrically around a central axis (cyclic homo-oligomers) are common in nature. This is not surprising if you consider that it’s more efficient to assemble something from uniform building blocks rather than irregular ones, and that symmetrical structures provide stability and definition (i.e., aggregation stops at a defined number of subunits). Cyclic homo-oligomers play key roles in many biological processes, including cell signaling, enzymatic catalysis, and protein function. Nevertheless, there has been no systematic approach to designing them starting from a monomeric protein structure.
Researchers in Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging Division worked with University of Washington’s David Baker and his team to design and crystallize self-assembling cyclic homo-oligomer proteins. A strategy was developed to design interfaces onto idealized proteins aimed to direct their assembly into multimeric complexes. The researchers used structural characterization at the ALS—both x-ray crystallography at Beamline 5.0.2 and small-angle x-ray scattering at Beamline 12.3.1—to show that many of the designs adopted the target oligomerization state and predicted structure. Not only does their work demonstrate that scientists have a basic understanding of what determines oligomerization, it also shows that they can design proteins with tunable shape, size, and symmetry for a variety of biological applications.
By predicting and designing these higher-order oligomers, they have gained an understanding of the fundamental principles underlying oligomer–oligomer interactions. Beyond the internal interactions of the protein itself, these designed oligomers can be used to explore basic questions about how the structure of signaling molecules affects the behavior of receptors and cellular response.
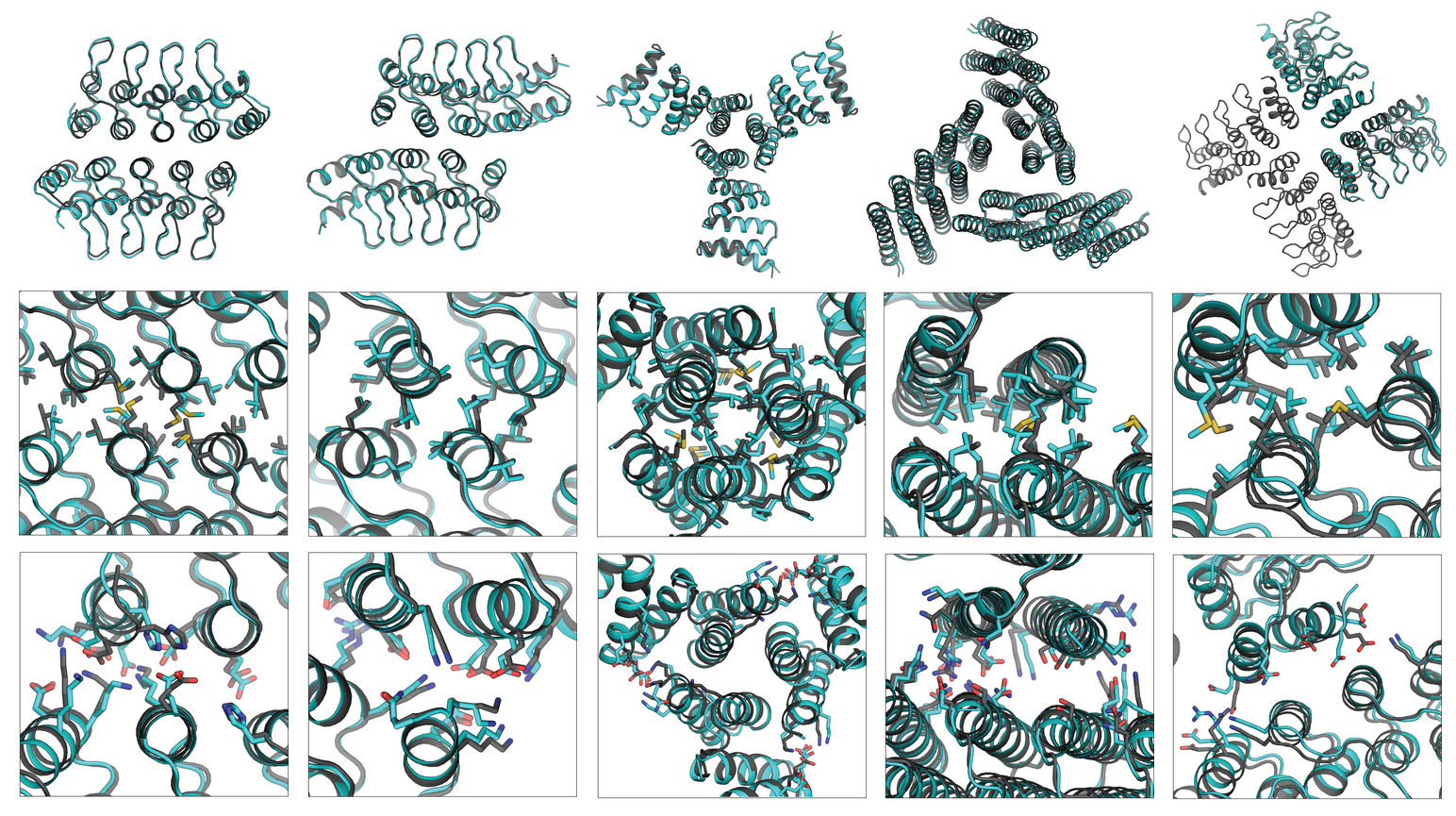
Work performed at ALS Beamlines 5.0.2 and 12.3.1.
J.A. Fallas, G. Ueda, W. Sheffler, V. Nguyen, D.E. McNamara, B. Sankaran, J.H. Pereira, F. Parmeggiani, T.J. Brunette, D. Cascio, T.R. Yeates, P. Zwart, and D. Baker, “Computational design of self-assembling cyclic protein homo-oligomers,” Nat. Chem. 9, 353 (2017), doi:10.1038/nchem.2673.
Adapted from the Berkeley Lab Science Short, “Designing Cyclic Oligomers: Greater Than the Sum of Their Parts.”