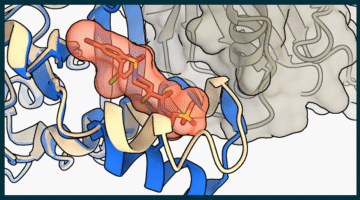
Researchers identified a compound that disrupts a hard-to-target tumor growth pathway in breast, lung, and other cancers and used the ALS to characterize the chemical interactions critical to its potency. This work contributed to the development of a similar compound currently undergoing clinical trials in cancer patients, and informs hypotheses for designing better drug candidates. Read more »
X-Rays Shed Light on Possible New Treatments for TB
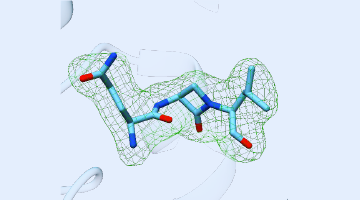
Using ALS beamlines, a new study revealed how CMX410 inhibits Pks13, a cell wall enzyme in Mycobacterium tuberculosis, the bacterium responsible for tuberculosis. CMX410 is effective against drug-sensitive and drug-resistant strains of the bacterium and has been proven safe in multiple animal models of infection. Read more »![]()
![]()
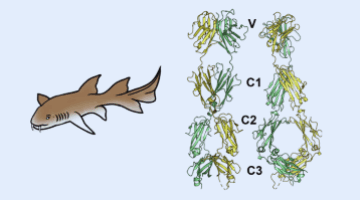
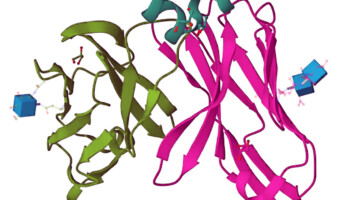
Sharks Shed Light on Origins of Adaptive Immune System
A team of researchers identified the three-dimensional structure of a protein expressed by a gene of a modern nurse shark that is proposed to be a close homologue to a gene that, more than 500 million years ago, gave rise to the adaptive immune system shared by all vertebrates. By understanding the emergence and evolution of the immune system, researchers may advance work in immunology, genetics, and biotechnology.
Read more »![]()
![]()
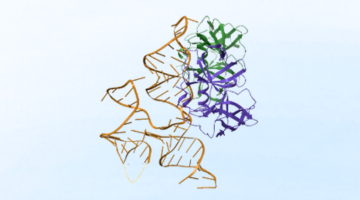
Researchers Identify Viral Swiss Army Knife, Clarifying How Replication Occurs
Viruses are ingenious, infectious agents, capable of replicating inside the living cells of a host organism. Enterovirus, a common viral pathogen, is responsible for a range of diseases from mild colds to severe conditions, including viral meningitis, myocarditis, and paralysis. A new study sheds light on how enteroviruses use structured RNA elements and multifunctional proteins to coordinate viral replication efficiently using minimal genetic material. Read more »
ALS Captures Structure of Engineered Protein, Opening New Options to Treat IBD
Researchers use the ALS to confirm the structure of an engineered immune protein that could open new opportunities to treat inflammatory bowel disease. Read more »
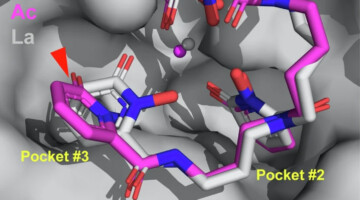
A Macromolecular Scaffold for Probing Actinium Chemistry
By encapsulating actinium atoms within a macromolecular complex for analysis using protein crystallography, researchers discovered that actinium has a unique solid-state bonding configuration. A better understanding of actinium behavior could help improve a promising cancer treatment known as targeted alpha therapy. Read more »![]()
![]()
Protein Pioneer: Enabling Scientists to Design Novel Proteins for the Future
The 2024 Nobel Prize in Chemistry was awarded to David Baker, Demis Hassabis, and John M. Jumper for the development of protein structure prediction and design. At the ALS, Baker leveraged high-throughput small-angle x-ray scattering (SAXS) and protein crystallography capabilities to design novel proteins and pave a new pathway for science, technology, and the environment. Read more »
Caught in the Actinium
In this work, researchers demonstrated a macromolecular scaffold that combines an 8-coordinate synthetic ligand and a mammalian protein to characterize the solution and solid-state behavior of the longest-lived actinium isotope. The information could help design better cancer treatments. Read more »
Computer-Aided Protein Design for New Biomaterials
Using a computer-based approach, researchers designed porous protein crystals that were revealed to be stable, tunable, and atomically accurate using x-ray scattering and diffraction at the ALS. The work provides a powerful new platform for biological materials engineering and opens up wide applications in biotechnology and medicine. Read more »![]()
![]()
Gemini Beamline 2.0.1 Banks Its First Protein Structure
A protein structure obtained from ALS Beamline 2.0.1 (“Gemini”) has recently been published in the literature and deposited into the Protein Data Bank (PDB)—two significant firsts for this beamline. The structure helped provide new insights into the molecular mechanisms involved in triggering certain inflammatory diseases. Read more »
- 1
- 2
- 3
- …
- 10
- Next Page »