SCIENTIFIC ACHIEVEMENT
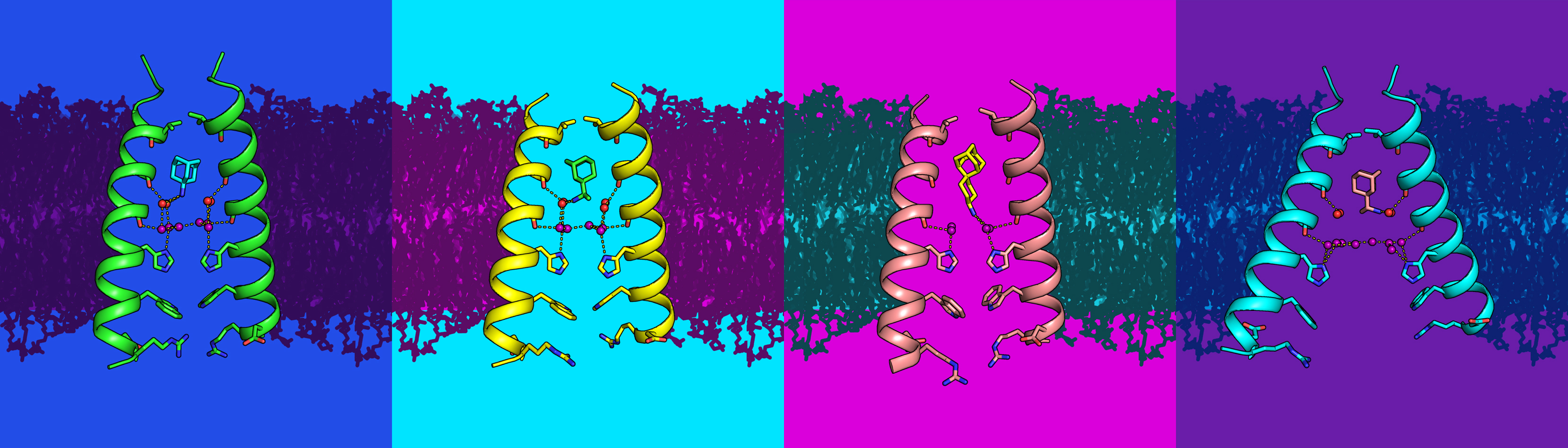
Researchers obtained high-resolution structures of several influenza antiviral drug molecules bound to their proton-channel targets in both open and closed conformations.
SIGNIFICANCE AND IMPACT
The structures provide an atomic-level blueprint from which to design more effective anti-influenza drugs that can overcome growing drug resistance.
Flu treatment goes (anti-)viral
Influenza virus infection is a perennial problem. According to the Centers for Disease Control and Prevention, the 2017-18 flu season saw high levels of emergency-department visits for influenza-like illness, high influenza-related hospitalization rates, and elevated and geographically widespread influenza activity for an extended period.
Although yearly vaccinations can reduce the number of flu infections, these vaccines are able to target only a subset of viral strains—there is, as yet, no “universal vaccine.” As a result, there is still a need for antiviral drugs to treat the illness after infection has occurred. This is especially important for groups of people who can experience serious complications from the flu, such as those with respiratory diseases or immune disorders. In recent years, however, resistance to certain classes of antiviral drugs has become a problem.
Adamantanes: diamondoid drugs
An adamantane molecule is a diamondoid—a few carbon atoms arranged in the diamond crystal structure, with hydrogen atoms on the surface. Some adamantane derivatives have been used in the treatment of influenza A because they were found to inhibit the function of membrane proteins (M2 proton channels) embedded in the viral envelope. Normally, as protons pass through these channels, the viral core becomes acidic, triggering the uncoating of viral RNA. Disruption of this channel thus blocks replication of the virus.
In recent years, however, resistance to M2-targeting antivirals has become widespread; most currently circulating influenza viruses are now resistant to adamantanes. The only orally bioavailable anti-influenza medications at present are not proton-channel blockers, but enzyme blockers (such as Tamiflu).
Previous structure-based studies have been used to inform the design of effective M2 inhibitors. But studies with resolution high enough to examine the critical role played by networks of hydrogen-bonded water molecules in the channel have been lacking. Such networks are widely utilized throughout nature to facilitate proton diffusion within proteins.
Cork in a bottle

At ALS Beamline 8.3.1, researchers have used protein crystallography to obtain high-resolution structures of wild-type (nonresistant) M2 transmembrane domains bound to two adamantane drug molecules (amantadine and rimantadine) and a novel M2 inhibitor (spiro-adamantyl amine). These complexes, in channel conformations both open and closed to the viral interior, provide the first high-resolution views of how the drugs interact with and disrupt the water-molecule networks lining the M2 channel.
Under normal function, M2 stabilizes hydronium (H3O+) at specific sites within the channel. The new structures reveal that ammonium groups on the drug molecules act as hydronium mimics, binding at sites that would normally attract hydronium. When this happens, the adamantane is positioned to block hydronium ions from reaching the channel pore, preventing protons from flowing through the channel like a cork stopping up a bottle.
Further experiments will involve characterizing the structure of drug-resistant M2 mutants and visualizing how new classes of inhibitors bind to them. Not only will this inform the design of new drugs to target adamantane-resistant mutations, it will also be relevant to the design of drugs that bind to the water-filled pores of channel proteins in general.
Contact: Jessica Thomaston
Researchers: J.L. Thomaston, N.F. Polizzi, and W.F. DeGrado (Univ. of California, San Francisco); A. Konstantinidi and A. Kolocouris (National and Kapodistrian University of Athens, Greece); and J. Wang (Univ. of Arizona).
Funding: National Institutes of Health and Univ. of California, Office of the President. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences Program (DOE BES).
Publication: J.L. Thomaston, N.F. Polizzi, A. Konstantinidi, J. Wang, A. Kolocouris, and W.F. DeGrado, “Inhibitors of the M2 proton channel engage and disrupt transmembrane networks of hydrogen-bonded waters,” J. Am. Chem. Soc. 140, 15219 (2018), doi:10.1021/jacs.8b06741.
ALS SCIENCE HIGHLIGHT #387