A powerful new tool for genome editing and gene regulation has emerged in the form of a family of enzymes known as Cas9. Cas9 could become an even more valuable tool with the creation of the first detailed picture of its three-dimensional shape. An international collaboration used x-ray crystallography to produce high-resolution structures of two major types of Cas9 enzymes. Combined with electron microscopy, the results point the way to the rational design of new and improved versions of Cas9 enzymes for basic research and genetic engineering.
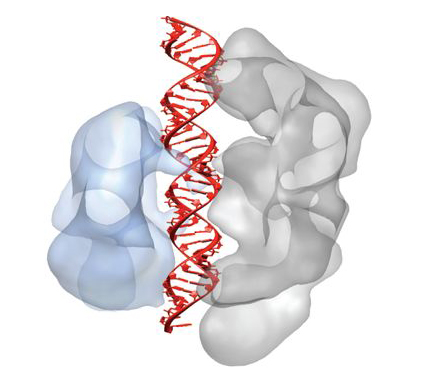
Cas9 enzymes play a key role in bacterial immune systems. Bacteria face a never-ending onslaught from viruses and invading strands of nucleic acid known as plasmids. To survive, they deploy a variety of defense mechanisms, including an adaptive immune system involving sequences in their DNA known as CRISPR (clustered regularly interspaced short palindromic repeats). A combination of CRISPR and squads of CRISPR-associated (Cas) proteins utilize small, customized RNA molecules (guide RNA) to target and cleave critical portions of an invader’s double-stranded DNA and confer immunity from similar invasions in the future.
The potential is there for bacteria and other microbes to be genetically engineered to perform valuable services, from the production of safer, more effective medicines and sustainable fuels, to the clean-up and restoration of our air, water, and land. Cells from more advanced organisms can also be modified for research or to fight disease. To achieve these and other worthy goals, the ability to precisely edit the instructions contained within a target’s genome is a must. Genetic engineers have already begun harnessing Cas9 for genome editing and gene regulation. However, despite successes to date, the technology has yet to reach its full potential because the structural basis for guide-RNA recognition and DNA targeting by Cas9 had been unknown.
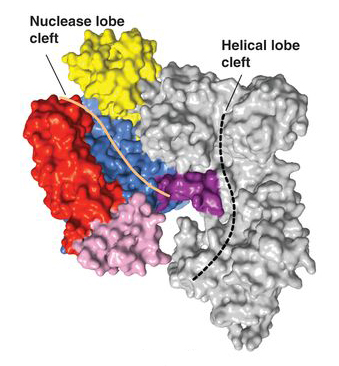
Now, researchers have addressed this knowledge deficit by first solving the three-dimensional crystal structures of two Cas9 proteins, representing large and small versions, from Streptococcus pyogenes (SpyCas) and Actinomyces naeslundii (AnaCas9) respectively. Using protein crystallography Beamlines 8.2.2 and 8.3.1 at the ALS and PXI and PXIII at the Swiss Light Source, the researchers discovered that despite significant differences outside of their catalytic domains, all members of the Cas9 family share the same structural core. The high-resolution (2.6- and 2.2-Å) images showed this core to feature a clam-shaped architecture with two major lobes—a nuclease domain lobe and an alpha-helical lobe. Both contain conserved clefts that become functional in nucleic-acid binding.
The researchers also employed electron microscopy to visualize the Cas9 bound to either guide RNA, or both RNA and target DNA. The images revealed that the guide RNA structurally activates Cas9 by creating, between the two main lobes, a channel that functions as the DNA-binding interface. The results underline that, in addition to sequence complementarity, other features of the guide RNA must be considered when employing this technology. The Cas9 protein, on its own, exists in an inactive state, but upon binding to the guide RNA, the Cas9 protein undergoes a radical change in its three-dimensional structure that enables it to engage with the target DNA.
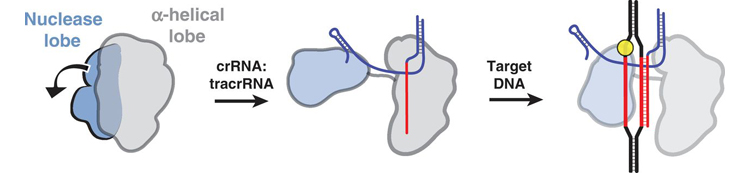
With these high-resolution structures of the two major types of Cas9 proteins, researchers can start to see how this family of bacterial enzymes has evolved. The two structures are quite different from each other outside of their catalytic domains, suggesting an interesting structural plasticity that could explain how Cas9 is able to use different kinds of guide RNAs. Also, the differences in the two structures suggest that it may be possible to engineer smaller Cas9 variants and still retain function, an important goal for some genome-engineering applications.
Contacts: Jennifer Doudna and Eva Nogales
Research conducted by: M. Jinek and C. Anders (University of Zurich); F. Jiang, S.H. Sternberg, E. Kaya, E. Ma, M. Hauer, and A.T. Iavarone (Univ. of California, Berkeley); D.W. Taylor, K. Zhou, and S. Lin (Howard Hughes Medical Institute and UC Berkeley); M. Kaplan (HHMI); E. Charpentier (Umeå University, Sweden; Helmholtz Centre for Infection Research, Germany; and Hannover Medical School, Germany); and E. Nogales and J.A. Doudna (UC Berkeley, HHMI, and Berkeley Lab).
Research funding: Howard Hughes Medical Institute, Bill and Melinda Gates Foundation, National Science Foundation, University of Zurich, European Research Council, China Scholarship Council, German Academic Exchange Program, National Defense Science and Engineering Graduate Fellowship, and Swedish Foundation for International Cooperation in Research and Education. Operation of the ALS is supported by the U.S. Department of Energy, Office of Basic Energy Sciences.
Publication about this research: M. Jinek, F. Jiang, D.W. Taylor, S.H. Sternberg, E. Kaya, E. Ma, C. Anders, M. Hauer, K. Zhou, S. Lin, M. Kaplan, A.T. Iavarone, E. Charpentier, E. Nogales, and J.A. Doudna, “Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation,” Science 343, 1247997 (2014).
ALS SCIENCE HIGHLIGHT #293