During the last two years, there has been an ever-increasing need for methods to rapidly analyze virus-infected cells in 3D and at high resolution. To respond in a relatively short time frame, scientists had to both collect and validate large amounts of data. For example, to understand the impact of SARS-CoV-2 at the cellular level, a group of scientists led by Carolyn Larabell at Berkeley Lab, in collaboration with investigators at the University of Heidelberg, safely visualized COVID-infected lung cells and identified the major hallmarks of infection.
The researchers collected and analyzed 17 individual cells using soft x-ray tomography (SXT) at Advanced Light Source Beamline 2.1. “The methodology is quantitative,” said Larabell, “and allows the imaging of whole cells in three dimensions without the need for chemical stains or the generation of multiple thin (80–100 nm) plastic sections.”
In SXT, cells typically are immobilized by rapid plunge-freezing, avoiding the need for chemical fixatives. However, to safely handle cells infected with highly contagious viruses, Larabell and collaborators established an aldehyde-based protocol to neutralize the infected cells prior to their arrival. “Many infectious organisms require a biosafety level 3 facility to be safely manipulated,” Larabell said, “but we managed to develop a way to study highly pathogenic agents with SXT in a BSL-2 approved laboratory.”
The technique uses the naturally occurring differential absorption of carbon-rich components in the “water window” energy range (283–580 eV). The absorption in this range is informative about the concentration of cellular biomolecules and can also be used to identify and interpret organelles based on composition.
Thus, SXT enables analysis of organelle remodeling at high resolution (about 50 nm), unveiling hallmarks of SARS-CoV-2 infection. The researchers examined the large autophagosomes or autolysosomes involved in disposal of the viral replication machinery, the double-membraned vesicles and convoluted membranes that enable virus replication, and multi-nucleated cells implicated in virus spreading. Amazingly, the technique requires only 10 minutes to collect and reconstruct an entire cell, increasing the number of samples that can be investigated in a single day and enabling scientists to respond to the increased demand for new data in a relatively short time frame.
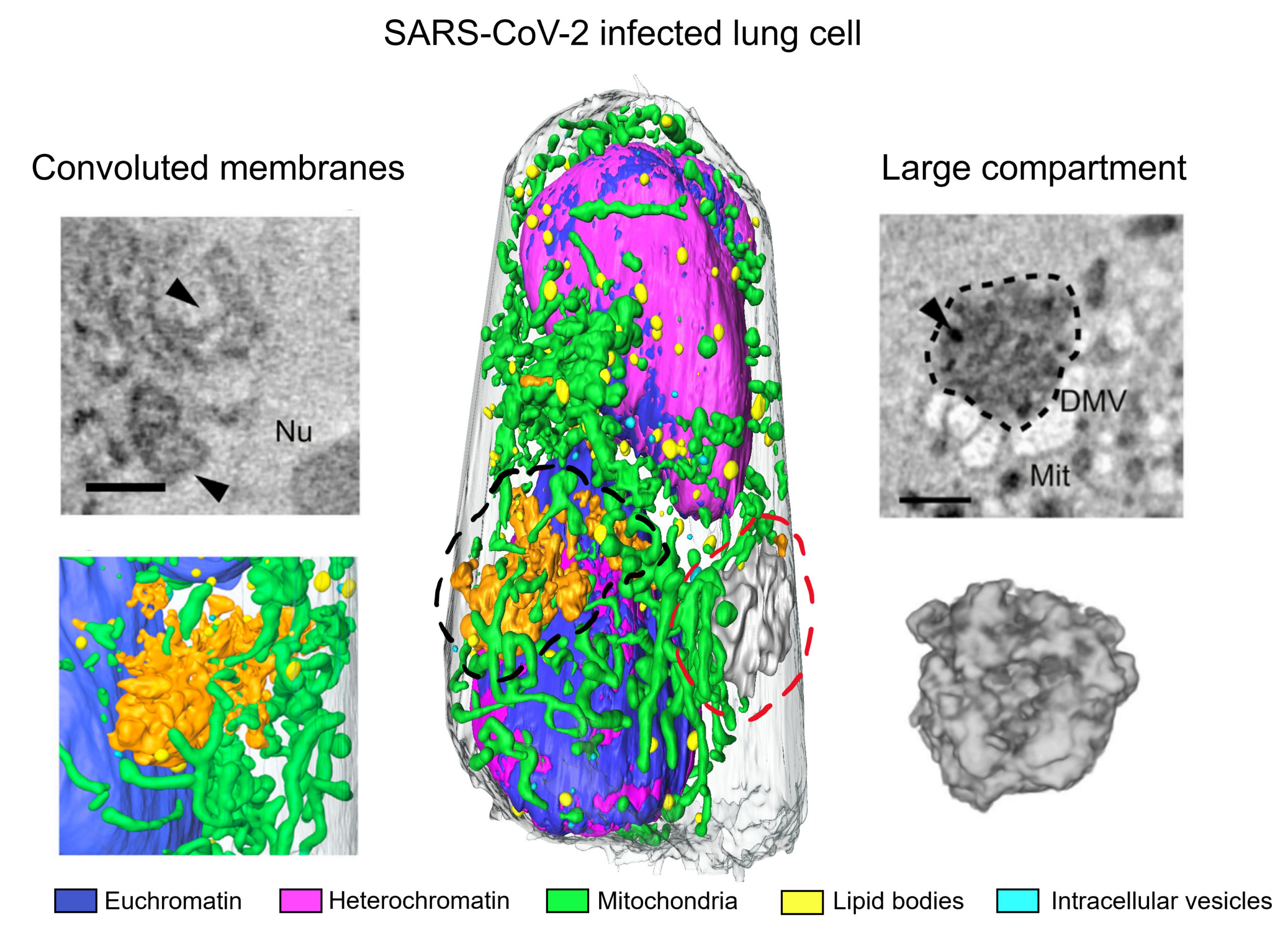
V. Loconte, J.-H. Chen, M. Cortese, A.A. Ekman, M.A. Le Gros, C.A. Larabell, R. Bartenschlager, and V. Weinhardt, “Rapid whole-cell quantitative imaging of structural changes induced by SARS-CoV-2 using soft x-ray tomography,” Cell Rep. Methods 1, 100117 (2021), doi:10.1016/j.crmeth.2021.100117.