by Lori Tamura
The eight structural biology beamlines at the ALS have now collectively deposited over 7000 proteins into the Protein Data Bank (PDB), a worldwide, open-access repository of protein structures. The 7000th ALS protein structure (entry no. 6C7C) is an enzyme from Mycobacterium ulcerans (strain Agy99), solved with data from Beamline 5.0.2. This bacterium produces a toxin that eats away at skin tissue, causing what’s known as Buruli ulcers (Google at your own risk!). The bacterium is antibiotic-resistant, and treatment involves the surgical removal of infected tissues, including amputation.
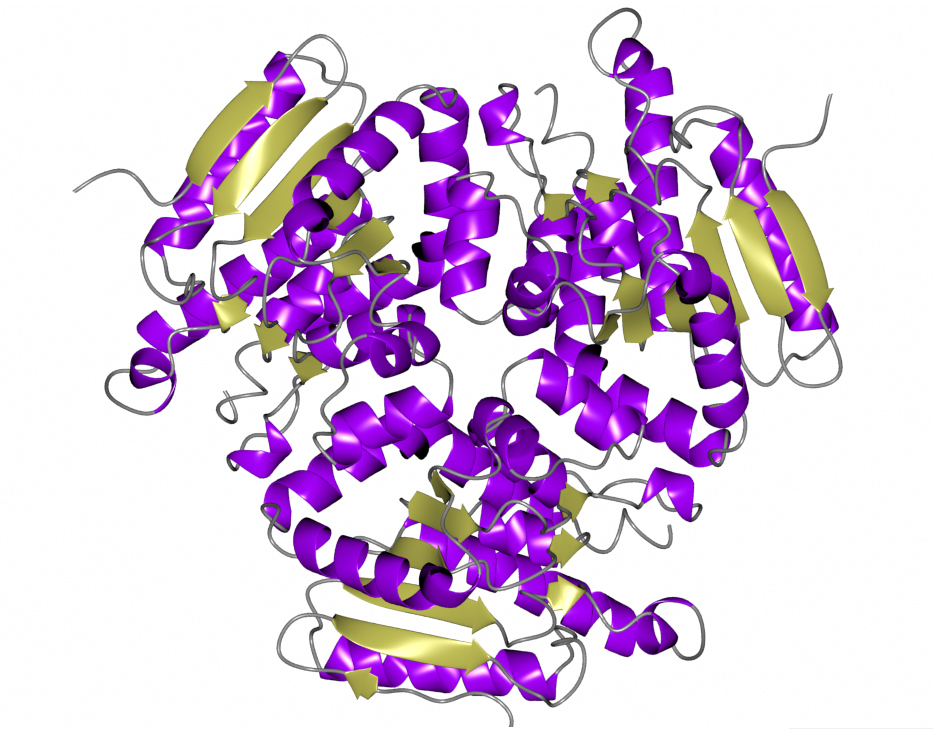
The enzyme structure was solved by a group from the Seattle Structural Genomics Center for Infectious Disease (SSGID), whose mission is to obtain crystal structures of potential drug targets on the priority pathogen list of the National Institute of Allergy and Infectious Diseases (NIAID). As of May 2018, SSGCID has deposited 1090 structures in the PDB, with data for more than a quarter of those collected at ALS beamlines.
The specific enzyme structure they solved is an Enoyl-CoA hydratase, which catalyzes the breakdown of fatty acids for the production of energy. The focus on this protein is part of the group’s “homolog rescue” strategy: when progress on a target protein suffers from low expression levels or challenging crystallization, they improve their chances of finding a viable drug target by looking at related species (homologs) with conserved active sites. Thus, information about M. ulcerans could also apply to M. tuberculosis, a related bacterium responsible for one of the world’s deadliest diseases.
The first protein crystallography beamline at the ALS, Beamline 5.0.2 came online in 1997, with a “wiggler” insertion device as its source of hard x-rays. The first protein solved at the ALS was deposited in the PDB a year later, in 1998. Since then, the structural biology contingent has grown, with two more beamlines soon established at the wiggler (Beamlines 5.0.1 and 5.0.3), and superbend beamlines coming online starting in the early 2000s (Beamlines 4.2.2, 8.2.1, 8.2.2, 8.3.1, and 12.3.1).
SSGCID is a consortium of four Pacific Northwest organizations (Center for Infectious Disease Research, Beryllium Discovery, University of Washington, and Washington State University). It is funded by NIAID for the purpose of structurally characterizing infectious species and enabling development of new therapeutic strategies.
The PDB was established in 1971 at Brookhaven National Laboratory and originally contained 7 structures. Today, nearly 50 years later, it contains over 140,000 biological macromolecular structures, enabling breakthroughs in research and education.
