SCIENTIFIC ACHIEVEMENT
An antibody that appears to neutralize all known SARS-CoV-2 strains and closely related coronaviruses was discovered with the help of protein-structure data from the Advanced Light Source.
SIGNIFICANCE AND IMPACT
The work highlights principles underlying antibody potency, breadth, and escapability that can guide the development of therapeutics against the current and potential future pandemics.
Progress in COVID-19 treatments
Lifesaving COVID-19 vaccines are allowing us to feel optimistic again, after more than a year of anxiety and tragedy. But vaccines are only one side of the coin—we also need treatments that can prevent severe disease after someone has been infected. In the past year, there has been significant progress in developing effective antibody-based therapies, and three drugs are currently available through emergency use authorization (EUA) by the Food and Drug Administration.
Sotrovimab, the newest antibody therapy, was developed by GlaxoSmithKline and Vir Biotechnology after a large collaborative study by scientists from across the nation discovered a natural antibody (in the blood of a SARS survivor, back in 2003) that has remarkable breadth and efficacy.
Experiments showed that this antibody, called S309, neutralizes all known SARS-CoV-2 strains—including newly emerged mutants that can now “escape” from previous antibody therapies—as well as the closely related original SARS-CoV-2 virus.
Structural maps point the way
Jay Nix, leader of the Molecular Biology Consortium based at the ALS, used Beamline 4.2.2 at the ALS and SLAC’s Stanford Synchrotron Radiation Lightsource to perform protein crystallography on samples of survivor-derived antibodies during an early phase of the study. His work, alongside other crystallography and cryo-electron microscopy findings, helped generate detailed structural maps of how these antibodies bind to the SARS-CoV-2 spike protein, allowing the wider team to select the most promising contenders and advance them to cell culture- and animal-based studies. Following exciting lab results, the developers designed sotrovimab based on the structure of S309, and evaluated it in clinical trials.
The FDA granted an EUA for sotrovimab in late May after trials showed that people with mild to moderate COVID-19 infections who received an infusion of the therapy had an 85% reduction in rates of hospitalization or death, compared with placebo.
Follow-up study on viral escape
Understanding that new mutations could arise and that a novel pathogenic coronavirus could emerge from an animal–human crossover event, the scientists began a follow-up study to deeply explore what factors make antibodies resistant to viral escape and how certain antibodies are also broadly reactive against diverse, related viruses. Using biochemical and structural analysis, deep mutational scanning, and binding experiments, they identified one antibody with unparalleled universal potency.
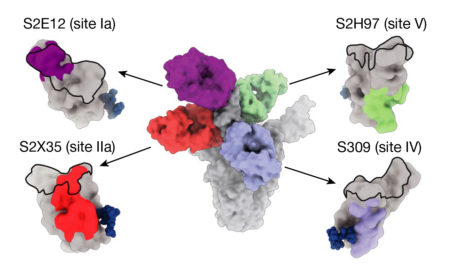
This antibody, which binds to a previously unknown site on the coronavirus spike protein, appears to neutralize all known sarbecoviruses—the genus of coronaviruses that cause respiratory infections in mammals. And, due to the unique binding site on a mutation-resistant part of the virus, it may well be more difficult for a new strain to escape. Subsequent tests in hamsters suggest that this antibody could even prevent a COVID-19 infection if given prophylactically.
In general, the work informs the development of antibody and vaccine countermeasures with greater robustness to immune escape in the current SARS-CoV-2 pandemic and utility for potential future sarbecovirus spillovers.
Contact: Tyler Starr
Researchers: T.N. Starr, A. Addetia, and A.S. Dingens (Fred Hutchinson Cancer Research Center); A.J. Greaney (Fred Hutchinson Cancer Research Center and University of Washington); N. Czudnochowski, P. Hernandez, J.A. Wojcechowskyj, L.E. Rosen, J. Zhou, M. Montiel-Ruiz, H. Kaiser, J.R. Dillen, H. Tucker, M.P. Housley, J. di Iulio, M. Agostini, M. Meury, E. Dellota Jr, C. Havenar-Daughton, A. Telenti, F.A. Lempp, C.M. Hebner, and G. Snell (Vir Biotechnology); Z. Liu, S. Stumpf, and S.P.J. Whelan (Washington University School of Medicine in St. Louis); F. Zatta, D. Pinto, M. Beltramello, R. Marzi, A. De Marco, J. Bassi, C. Silacci-Fregni, G. Lombardo, N. Sprugasci, K. Culap, S. Jaconi, E. Cameroni, L. Piccoli, F. Benigni, M.S. Pizzuto, and D. Corti (Humabs BioMed SA, Switzerland); Y.-J. Park, J.E. Bowen, M.A. Tortorici, A.C. Walls, and D. Veesler (University of Washington); W.G. Glass and J.D. Chodera (Memorial Sloan Kettering Cancer Center); I. Zhang (Memorial Sloan Kettering Cancer Center and Weill Cornell Graduate School of Medical Sciences); R. Abdelnabi, S.-Y.C. Foo,and J. Neyts (KU Leuven, Belgium); T.I. Croll (University of Cambridge); J.C. Nix (ALS); H.W. Virgin (Vir Biotechnology, Washington University School of Medicine in St. Louis, and University of Texas Southwestern Medical Center); and J.D. Bloom (Fred Hutchinson Cancer Research Center, University of Washington, and Howard Hughes Medical Institute).
Funding: Fred Hutchinson Cancer Research Center, National Institutes of Health, Arnold and Mabel Beckman Foundation, National Science Foundation, Damon Runyon Cancer Research Foundation, Gates Foundation, Pew Charitable Trusts, Burroughs Wellcome Fund, Wellcome Trust, Fast Grants, Bayer, Molecular Sciences Software Institute, Howard Hughes Medical Institute, and the U.S. Department of Energy (DOE) Office of Science, Biological and Environmental Research (BER) and Basic Energy Sciences (BES) programs. Operation of the ALS is supported by DOE BES.
Publication: T.N. Starr, N. Czudnochowski, Z. Liu, et al., “SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape,” Nature 597, 97 (2021), doi:10.1038/s41586-021-03807-6.
Adapted from the Berkeley Lab Snapshot, “Scientist at Berkeley Lab Played a Hand in ‘Inescapable’ COVID-19 Antibody Discovery.”
ALS SCIENCE HIGHLIGHT #454