SCIENTIFIC ACHIEVEMENT
Using x-ray crystallography performed in part at the Advanced Light Source (ALS), researchers studied 36 DNA-based molecular junctions and discovered factors that yield superior 3D self-assembled lattice structures.
SIGNIFICANCE AND IMPACT
The work expands the set of building blocks for lattices that can scaffold molecules into regular arrays, from proteins for structure studies to nanoparticles for nano-antennas and single-particle sensors.
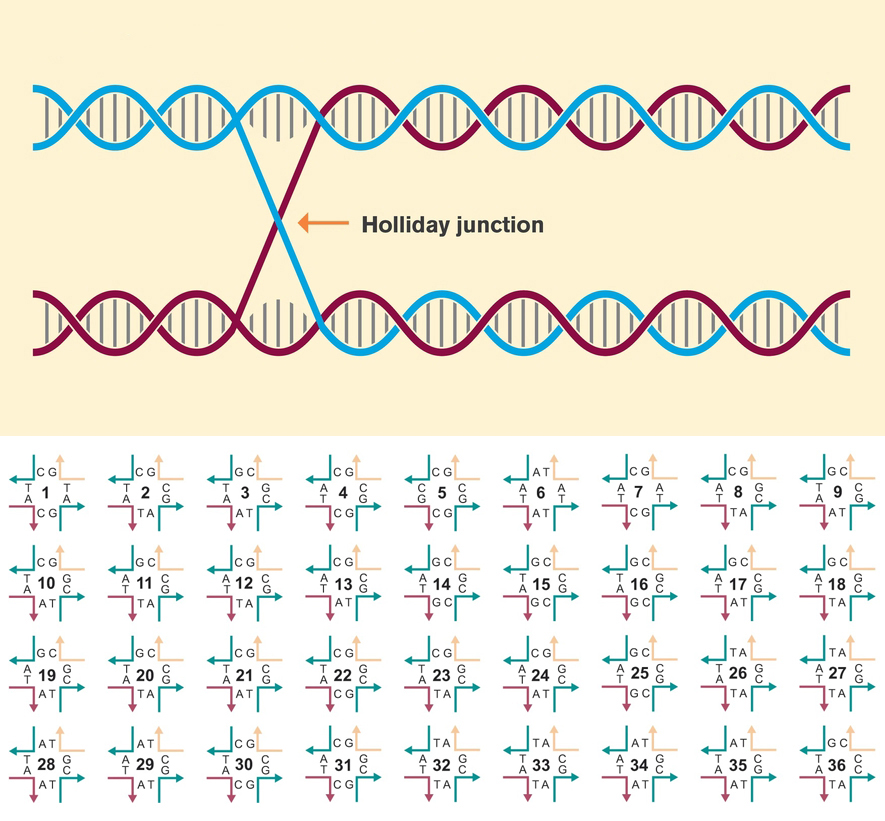
Holliday junction functions
Nature uses DNA’s double helix to store the blueprints of all living forms, drawing on a four-letter alphabet of nucleotides. Researchers in the field of DNA nanotechnology seek to emulate and even extend the possibilities of DNA architecture beyond what nature has created. One basic building block used in the fabrication of many DNA nanoforms is known as a Holliday junction. This nexus of two segments of double-stranded DNA has been used to form elaborate, self-assembling crystal lattices at the nanometer scale.
In this work, researchers studied the characteristics of 36 variants of the Holliday junction. They found that the effectiveness of a given variant for constructing crystalline nanoarchitectures is sensitively dependent not only on the arrangement of the four nucleotide pairs forming the junction, but also on sequences forming the junction’s four “arms” and the presence of ions captured within them. The resulting expanded set of DNA building blocks enables the precise arrangement of molecular components at the nanoscale.

A multipurpose crystal scaffold
The foundational goal of structural DNA nanotechnology is to rationally design self-assembling 3D crystals with DNA junctions. These scaffolds could one day enable scientists to position proteins, peptides, small molecules, and nanoparticles into regular arrays, bypassing standard crystallization screening methodologies for a host of target molecules that could not otherwise be crystallized for structural studies at atomic resolution. These DNA scaffolds could also be used as molecular sponges that “absorb” targets, whether for the site-specific positioning of proteins involved in enzyme cascades or to orient various nanoparticles for nanoelectronics or nanophotonics applications (e.g. nanoantennas, metamaterials, and single-molecule sensing).
Systematic structure studies
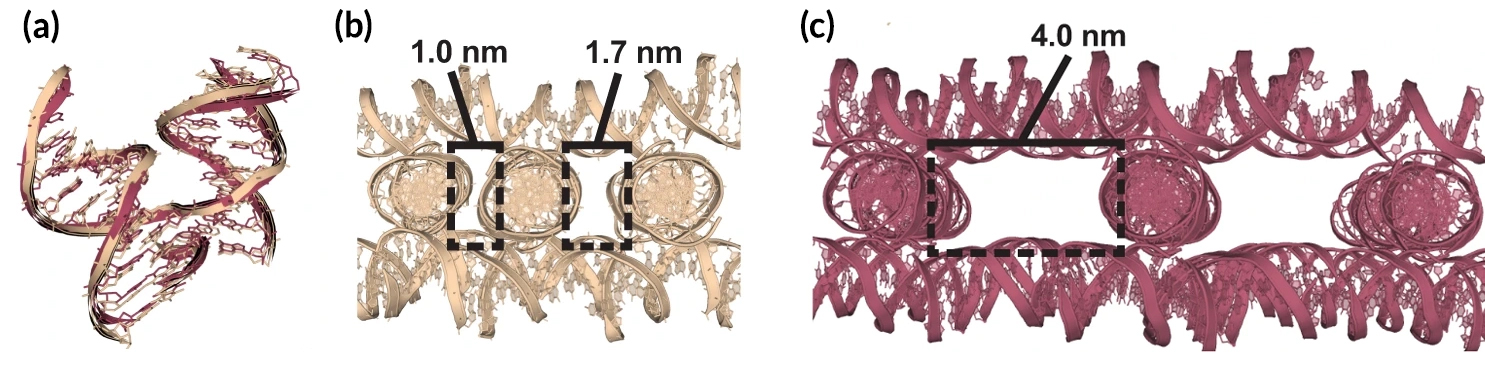
Here, researchers conducted the first systematic study of Holliday junctions for self-assembled DNA crystals using x-ray crystallography at Berkeley Center for Structural Biology (BCSB) Beamline 5.0.2 at the ALS, as well as at the Advanced Photon Source and the National Synchrotron Light Source II. Crystallography experiments with self-assembling DNA crystals have been conducted at BCSB beamlines since 2012, yielding several novel DNA crystal scaffolds that provided the foundation for the work described here. The current comprehensive study determined 134 unique crystal structures, about half of which (68) were solved directly from data obtained at the ALS. The sheer volume of the data collected required years of allocated beam time at BCSB beamlines.
An expanded DNA toolkit
For decades, one Holliday-junction nucleotide sequence (“J1”) had been used for all self-assembling DNA crystal systems because it was initially described as the most energetically favorable. In this study, however, researchers identified a host of superior junctions that not only improve resolution but also control crystal symmetry. Unexpectedly, it turns out that nucleotides adjacent to the junction can have a significant effect on lattice architecture as well as on resolution. In addition, six junction sequences were found to be completely resistant to crystallization. Molecular dynamics simulations revealed that these junctions invariably lack two ion-binding sites that are pivotal for crystal formation.
Efforts are currently underway to rationally design new crystal systems with varying symmetries for hosting guest materials. More broadly, the knowledge gained from this work expands the toolkit for the molecular engineering of DNA-based platforms for applied nanoelectronics, nanophotonics, and catalysis.
Contact: Chad Simmons
Researchers: C.R. Simmons, T. MacCulloch, M. Matthies, A. Buchberger, I. Crawford, P. Šulc, N. Stephanopoulos, and H. Yan (Arizona State University), and M. Krepl and J. Šponer (Institute of Biophysics of the Czech Academy of Sciences and Palacky University Olomouc, Czech Republic).
Funding: U.S. Department of Energy (DOE), Office of Science, Biological and Environmental Research program; Howard Hughes Medical Institute; National Institutes of Health; National Science Foundation; European Regional Development Fund; Czech Science Foundation; and Arizona State University. Operation of the ALS is supported by DOE, Office of Science, Basic Energy Sciences program.
Publication: C.R. Simmons, T. MacCulloch, M. Krepl, M. Matthies, A. Buchberger, I. Crawford, J. Šponer, P. Šulc, N. Stephanopoulos, and H. Yan, “The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly,” Nat. Commun. 13, 3112 (2022), doi:10.1038/s41467-022-30779-6
Adapted from the Berkeley Lab Biosciences Area highlight, “Study Expands Horizons for DNA Nanotechnology.”
ALS SCIENCE HIGHLIGHT #470