SCIENTIFIC ACHIEVEMENT
Using structural data from the Advanced Light Source (ALS) and cryo-electron microscopy, researchers have characterized how an antibody binds to and neutralizes SARS-CoV-2.
SIGNIFICANCE AND IMPACT
This work provides the basis for therapeutic and vaccine development for the SARS-CoV-2 virus, which is responsible for the COVID-19 pandemic.
Leveraging the immune system
As scientists across the globe race to develop a vaccine against SARS-CoV-2, the coronavirus that causes COVID-19, an international team led by Davide Corti at Vir Biotechnology and David Veesler at the University of Washington has been working around the clock on a complementary approach—identifying neutralizing antibodies that could be used as a preventative treatment or as a post-exposure therapy.
Neutralizing antibodies are small proteins that inhibit pathogens by binding to the molecule or molecules that the microbe or virus uses to infect host cells. In humans and other animals, special immune cells produce neutralizing antibodies in response to infections, so that if the same pathogen is encountered again, the body can eliminate it more quickly.
Though natural neutralizing antibodies are typically only produced in the body for a limited time after the initial infection—past research with coronaviruses shows neutralizing antibodies last one or two years—scientists can manufacture pharmaceutical quantities of identical antibodies as long as they know the protein sequence. Mass-produced antibodies may then be given to people who do not yet have any of their own antibodies against that particular pathogen.
Vaccines, on the other hand, induce the body to produce its own antibodies by introducing a carefully chosen part of a pathogen—typically a molecule from its outer surface, or a weakened or inert version of the entire pathogen.
SARS and MERS antibodies
Soon after SARS-CoV-2 emerged, Veesler and colleagues began screening for potential neutralizing antibodies among those identified from SARS and MERS survivors in 2003 and 2013, respectively. Their previous research had revealed that some neutralizing antibodies produced in response to those diseases were also effective against closely related coronaviruses.
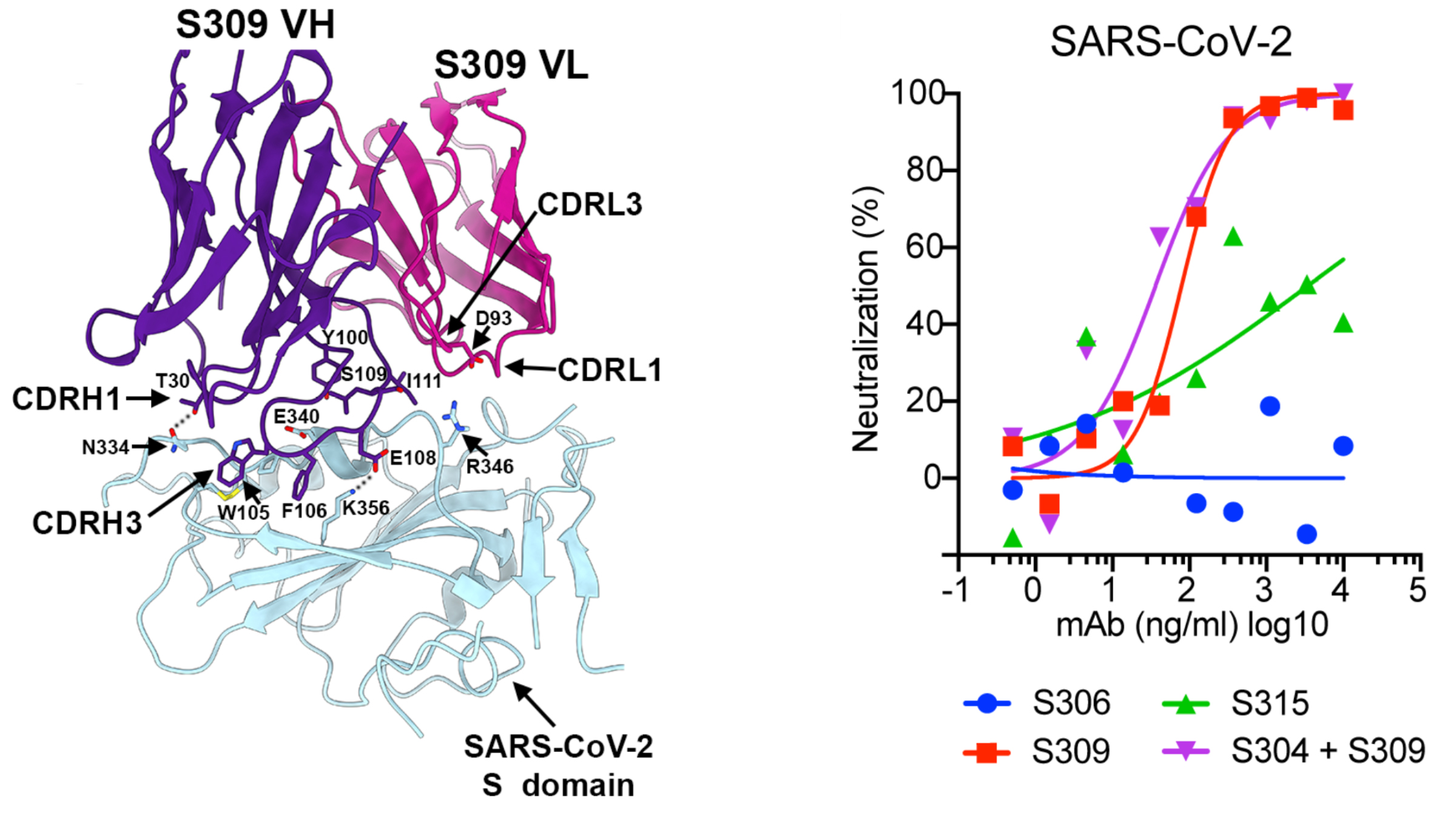
The screening yielded eight antibodies that can bind to the SARS-CoV-2 spike glycoprotein—a pyramid-shaped structure on the viral surface that facilitates entry into a host cell. Further tests narrowed the field to reveal one SARS-CoV antibody, S309, that successfully neutralizes SARS-CoV-2.
Mapping the structure
To understand how this antibody hinders the spike protein and to gather the information necessary to reproduce it, the team behind the current study used cryo-electron microscopy (cryo-EM) at the University of Washington and x-ray crystallography at ALS Beamline 5.0.2, managed by the Berkeley Center for Structural Biology (BCSB). Beamline 5.0.2 is a specialized crystallography beamline that has been in operation for 20 years, supporting a broad spectrum of structural biology studies and drug discovery.
In 2018, the Veesler Group had used the ALS to examine the spike glycoproteins of other coronaviruses and investigate how potential antibodies bind to them. When it became clear that SARS-CoV-2 was a threat, the ALS was able to give the team priority time to use the beamlines. The group used the ALS in early February, 2020, and on March 31, they analyzed crystallized samples of S309. The samples were not infectious and posed no safety risk.
Their overall findings, which include the structural data gathered at the ALS, indicate that antibodies derived from SARS survivors could potently block entry of SARS-CoV-2 and other closely related coronaviruses into host cells. The most promising candidate antibody is already on an accelerated development path toward clinical trials.
Contact: David Veesler
Researchers: D. Pinto, M. Beltramello, S. Bianchi, S. Jaconi, K. Culap, F. Zatta, A. De Marco, A. Peter, B. Guarino, E. Cameroni, K. Fink, and D. Corti (Humabs Biomed SA); Y.-J. Park, A.C. Walls, and D. Veesler (University of Washington); M.A. Tortorici (University of Washington and Institut Pasteur); R. Spreafico, C. Havenar-Daughton, G. Snell, A. Telenti, and H.W. Virgin (Vir Biotechnology); A. Lanzavecchia (Humabs BioMed SA and Università della Svizzera italiana); J.B. Case, R.E. Chen, and M.S. Diamond (Washington University School of Medicine).
Funding: National Institutes of Health, Pew Biomedical Scholars, Burroughs Wellcome Fund, University of Washington, Institut Pasteur, Vir Biotechnology Inc., Humabs BioMed SA, and Moderna. Operation of the ALS to conduct this research was supported in part by the U.S. Department of Energy (DOE) National Virtual Biotechnology Laboratory, a consortium of DOE National Laboratories with core capabilities relevant to the threats posed by COVID-19, and funded under the Coronavirus Aid, Relief, and Economic Security (CARES) Act. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences program.
Publication: D. Pinto, Y.-J. Park, M. Beltramello, A.C. Walls, M.A. Tortorici, S. Biachi, S. Jaconi, K. Culap, F. Zatta, A. De Marco, A. Peter, B. Guarino, R. Spreafico, E. Cameroni, J.B. Case, R.E. Chen, C. Havenar-Daughton, G. Snell, A. Telenti, H.W. Virgin, A. Lanzavecchia, M.S. Diamond, K. Fink, D. Veesler, and D. Corti, “Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody,” Nature 583, 290 (2020), doi:10.1038/s41586-020-2349-y.
Adapted from the Berkeley Lab news release, “X-ray Experiments Zero in on COVID-19 Antibodies.”
ALS SCIENCE HIGHLIGHT #422