“Viral factories” are areas in virus-infected host cells where the tools and materials necessary for viral replication are concentrated. In this study, researchers sought to learn more about an important component of some viral factories, a protein called σNS. This protein takes part in the replication of reoviruses, which are generally nonpathogenic and can be used as an oncolytic agent to target cancer cells. Despite its importance, the underlying mechanics of σNS have remained unclear.
A collaborative team led by B.V.V. Prasad at Baylor College of Medicine and Terence Dermody at the University of Pittsburgh conducted protein crystallography studies at Beamline 5.0.1 of the Advanced Light Source. They looked at a mutant version of σNS, σNS-R6A, which forms dimers rather than the longer chains (oligomers) of the unmutated protein, which resists crystallization.
The team discovered that σNS-R6A dimers interact by inserting protruding arms into a pocket of its neighbor, forming a helical assembly. The interior of the helical assembly is positively charged, making it suitable for binding RNA.
Bile acids were found to disrupt σNS assembly by binding the same pocket. “This was a serendipitous discovery,” said Prasad. “First author Boyang Zhao, a graduate student at the time, had set up crystallization trials with additives that included bile acid salts. When the crystal structure was determined, he saw bile acid moiety in the structure.”
The structure with bile acid revealed that the displaced arm, which is located at the N-terminal end of the protein, becomes flexible. The observations, together with structural and biochemical studies of σNS lacking the N-terminal arm, suggest that it transitions from a flexible state to an ordered structure to form σNS oligomers.
The researchers observed that σNS exhibits RNA-chaperone activity likely essential for presenting messenger RNA to the viral polymerase for replication. This activity is reduced by bile acids and abolished by N-terminal arm deletion, suggesting that the activity requires formation of σNS oligomers.
Overall, this research provides structural and mechanistic insights into the function of the σNS protein in reovirus replication. Understanding these mechanisms will foster development of therapeutic strategies against viruses that use σNS-like proteins to replicate.
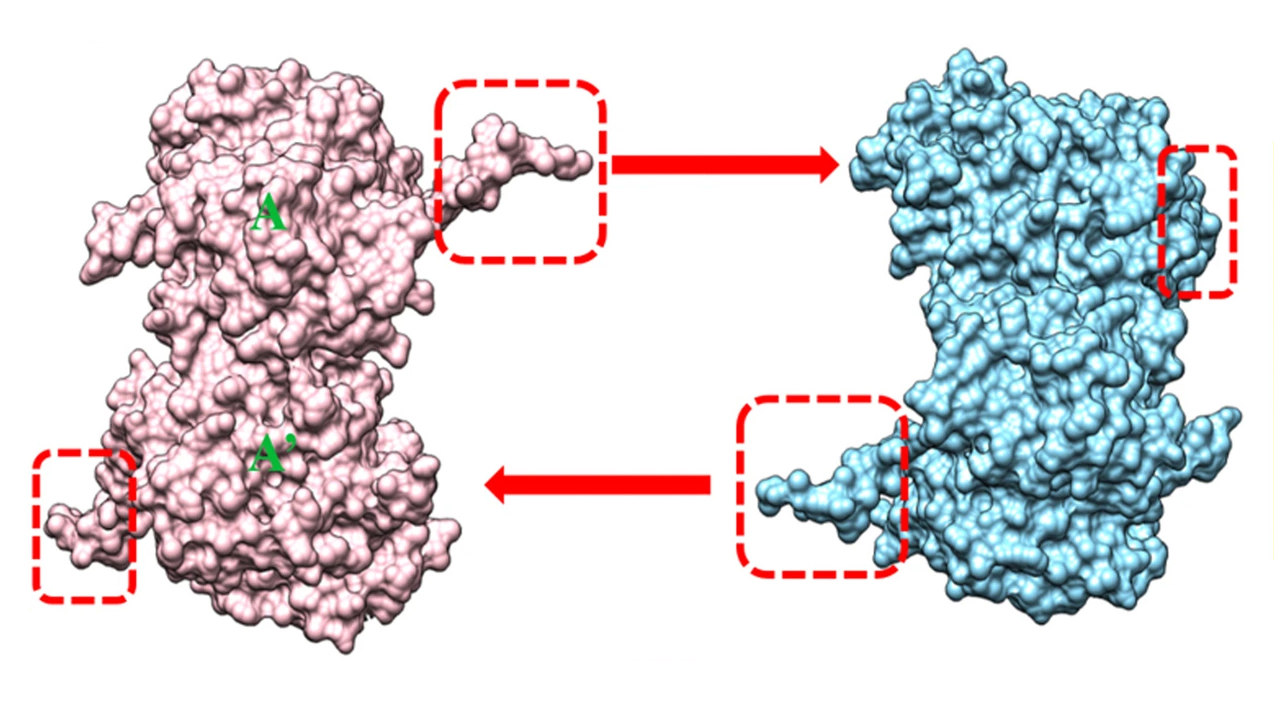
B. Zhao, L. Hu, S. Kaundal, N. Neetu, C.H. Lee, X. Somoulay, B. Sankaran, G.M. Taylor, T.S. Dermody, and B.V. Venkataram Prasad, “Structure of orthoreovirus RNA chaperone σNS, a component of viral replication factories,” Nat. Commun. 15, 2460 (2024), doi:10.1038/s41467-024-46627-8.