Viruses have evolved a wide variety of ways to exploit parts of their host cells to avoid detection and to propagate. For example, the hepatitis C virus (HCV), which attacks the liver, is known to repurpose host-cell components known as microRNAs—short RNA strands that act to silence gene expression. HCV harbors binding sites in its RNA genome for the liver-abundant microRNA, miR-122.
Surprisingly, previous work showed that, rather than destabilizing the viral RNA, as occurs with endogenous (host) RNA, miR-122 stabilizes the HCV genome. Further studies provided evidence that a site at the very end of the viral genome (Site-1) protects the uncapped end of the viral RNA from host antiviral responses and degradation by enzymes.
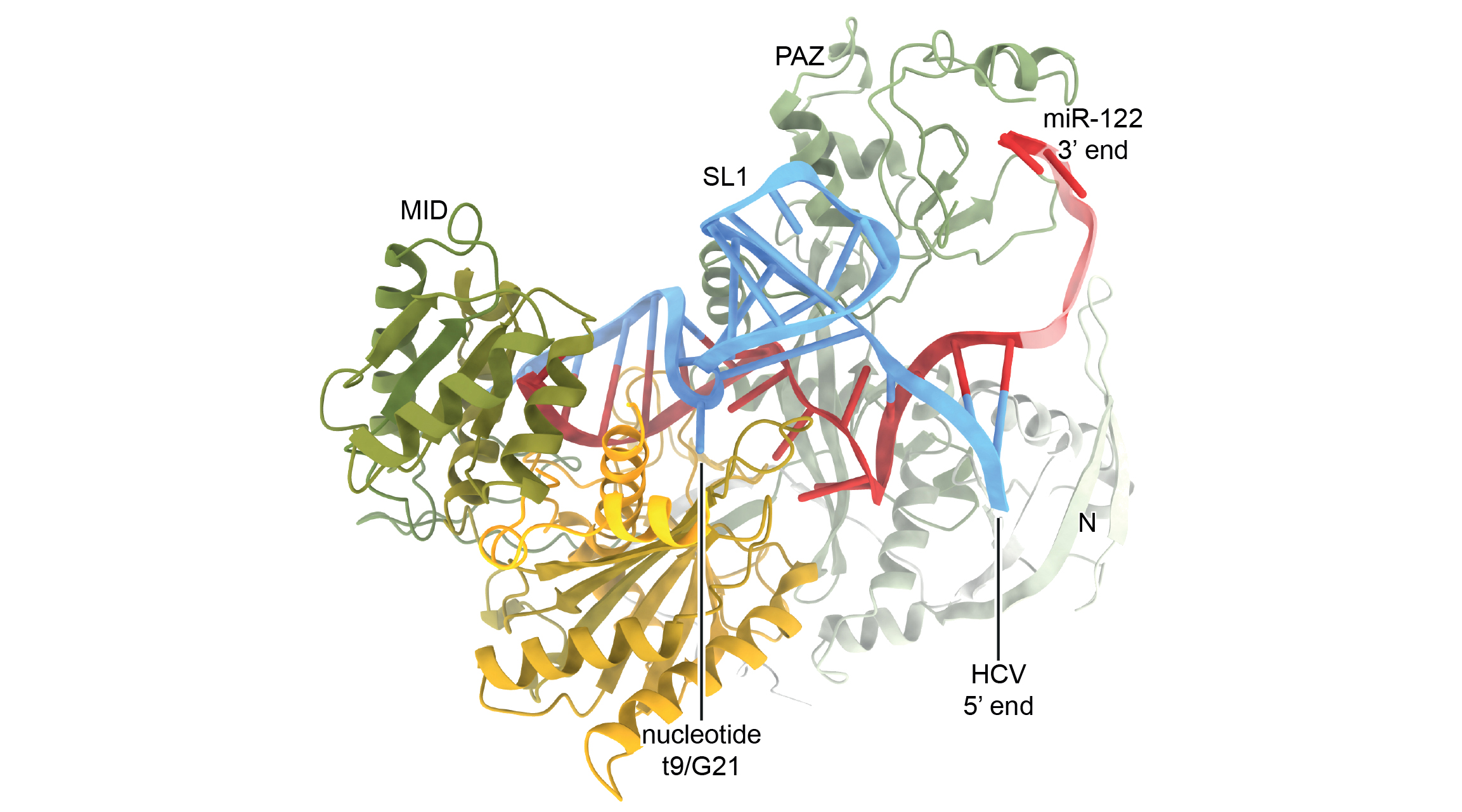
“Unlike known endogenous microRNA binding sites, Site-1 contains a conserved stem-loop, or hairpin structure, connecting regions where it interacts with miR-122,” said Luca Gebert, a scientist at the Scripps Research Institute in La Jolla. “So we were curious as to how this RNA structure interacted with the miR-122 complex, and whether it had any special significance.”
To determine the molecular mechanism of the viral microRNA binding site, Gebert and colleagues performed an array of biochemical and cell-based assays combined with x-ray crystallography at Advanced Light Source Beamline 5.0.2, where they obtained the structure of HCV Site-1 bound to miR-122 and Ago2, a closely associated protein that also plays a central role in RNA silencing.
The structure revealed how the viral RNA forms a functional motif that imparts a wide-open conformation onto Ago2, counteracting the natural propensity of the Ago2:miR-122 complex to laterally diffuse along the RNA, which would be detrimental to the virus. Instead, the viral RNA locks the complex in place at the uncapped end, greatly increasing protection from the cell’s antiviral response.
So far, the molecular properties of the peculiar miR-122 binding site appear unique, although superficially similar motifs have been observed in related viruses, including Zika. This leads the researchers to hypothesize that the motifs are evolutionary echoes of special endogenous microRNA target sites that have so far escaped detection, and perhaps enable microRNAs to serve in roles beyond RNA silencing.
L.F.R. Gebert, M. Law, and I.J. MacRae, “A structured RNA motif locks Argonaute2:miR-122 onto the 5’ end of the HCV genome,” Nat. Commun. 12, 6836 (2021).