Scientists have revealed for the first time the molecular steps that turn on bacteria’s pathogenic genes. Using an array of high-powered x-ray imaging techniques at the ALS, the researchers showed that certain bacterial DNA-binding proteins, denoted HU, are involved in the physical twisting of the genetic strand and that its supercoiling can trigger the expression of genes that make a microbe invasive. The study could open up new avenues in the development of drugs to prevent or treat bacterial infection.
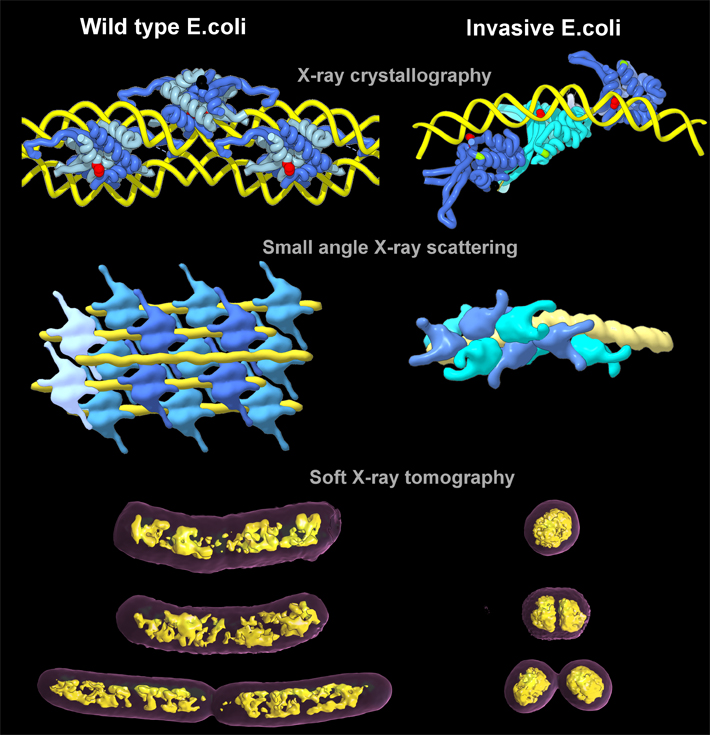
The researchers looked at how long strands of DNA are tightly packed into chromosomes, a necessity if they are to fit into compact spaces. In eukaryotes (organisms whose cells have a nucleus—including humans), the strands of DNA wrap around proteins called histones, like thread on a spool. In contrast, the DNA strands in single-celled prokaryotes (such as bacteria) are compacted into a region inside the cell called a nucleoid, analogous to a nucleus but not enclosed by a membrane. And like histone in eukaryotes, HU in prokaryotes helps organize the DNA into more-compact chromosomes.
However, when the normal twists and turns of DNA compaction turn into supercoiling (over- or under-winding of the double helix), trouble can begin. It has been known that DNA supercoiling leads to pathogenicity in bacteria, but exactly how the bacterial chromosome is condensed, organized, and ultimately segregated has been a puzzle for over half a century. In this work, the researchers were able to visualize for the first time how this packing is done in E. coli, and they also discovered that the way HU proteins pack the chromosomes can trigger gene expression.
Elucidating these molecular mechanisms entailed imaging interactions that span the nanoscale and mesoscale, from HU-DNA molecular complexes to bacterial chromosome and nucleoid structure, using three ALS beamlines. The Structurally Integrated Biology for Life Sciences (SIBYLS) Beamline 12.3.1 combines x-ray crystallography and small-angle x-ray scattering (SAXS) capabilities. The crystallography performed there, as well as at Beamline 8.3.1, provided atomic-level details of how the HU proteins—both wild type (normal) and invasive (pathogenic)—interacted with the bacterial DNA, while SAXS was able to show how the HU proteins assembled and affected the longer strands of DNA in solution.
Then, to get a clear sense of how that twisting and packing manifests at the cellular level, the team took their samples to the National Center for X-Ray Tomography (NCXT) at ALS Beamline 2.1. With x-ray tomography, they were able to see the natural contrast in organic material in as close to a living state as possible, and they obtained quantitative comparisons of how compacted the chromosomes were in normal and pathogenic strains of E. coli. The results showed that the genetic material in the pathogenic E. coli was so tightly packed that it consumed less than one-half the volume of its non-pathogenic counterpart.
Initially, it had been believed that the enzyme topoisomerase was the primary driver of DNA coiling in bacteria. This study showed that, independent of topoisomerase, changing the assembly of HU proteins was enough to induce changes in DNA coiling at different stages of bacterial growth. What is notable about HU proteins as a trigger for gene expression is that it is quick. This makes sense as a survival mechanism for bacteria, which need to adapt quickly to different environments. The study results also beg the question: If pathogenicity can be switched on, could it also be switched off? The researchers certainly expect to answer that question in future studies. These HU interactions could be an attractive target for drugs that control pathogenesis, not only of bacteria, but of other microbes with comparable genetic structures.

Contact: Michal Hammel
Research conducted by: M. Hammel, F.E. Reyes, R. Parpana, and H.Y.H. Tang (Berkeley Lab); C.A. Larabell (Berkeley Lab and UC San Francisco); J.-H. Chen (UC San Francisco); J.A. Tainer (Berkeley Lab and University of Texas MD Anderson Cancer Center); and D. Amlanjyoti and S. Adhya (National Institutes of Health).
Research funding: National Institutes of Health and the U.S. Department of Energy (DOE), Office of Biological and Environmental Research. Operation of the ALS is supported by the DOE Office of Basic Energy Sciences.
Publication about this research: M. Hammel, D. Amlanjyoti, F.E. Reyes, J.-H. Chen, R. Parpana, H.Y.H. Tang, C.A. Larabell, J.A. Tainer, and S. Adhya, “HU multimerization shift controls nucleoid compaction,” Sci. Adv. 2, e1600650 (2016). doi: 10.1126/sciadv.1600650
ALS SCIENCE HIGHLIGHT #338