Early in the COVID-19 pandemic, it was quickly established that the receptor binding domain (RBD) of the SARS-CoV-2 spike protein is a prime target for antibodies that neutralize the virus. Now, scientists have found that a second region of the spike protein, called the N-terminal domain (NTD), also contains a key “site of vulnerability” that is targeted by dozens of antibodies, some of which exhibit ultrapotent neutralizing activity. The findings were reported in a paper recently published in Cell.
“Knowing where neutralizing antibodies bind on the virus is critical in the design of vaccines and, as it turns out, essential to understanding the mutations that the virus is acquiring as it attempts to evade our immunity,” said the paper’s co-lead author, Matthew McCallum, a postdoctoral fellow in the University of Washington’s Department of Biochemistry.
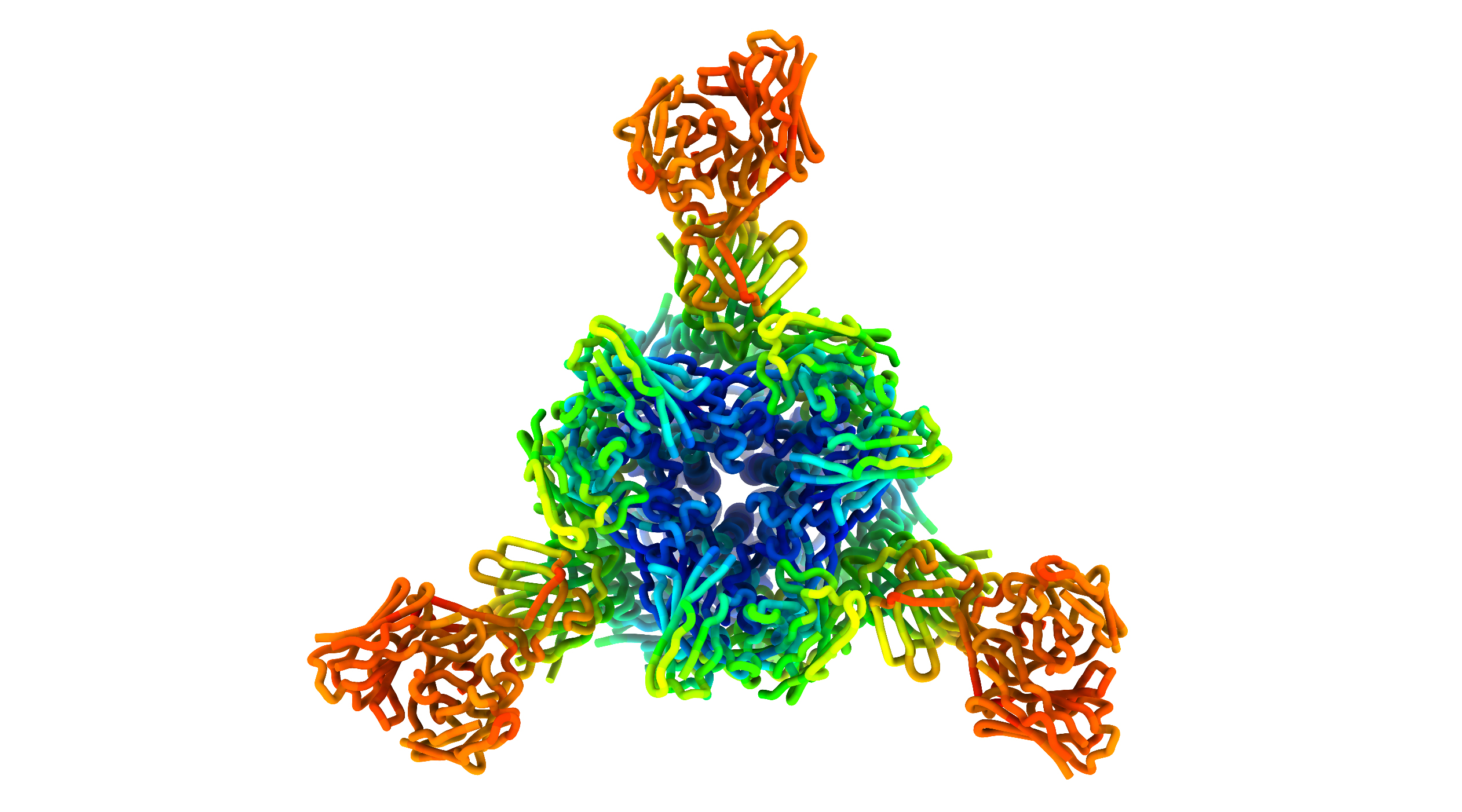
To understand the mechanisms behind the antibodies’ neutralizing ability, the researchers studied the way antibodies bind to the NTD and RBD regions of the spike protein, using cryogenic electron microscopy at the University of Washington as well as protein crystallography at Beamline 5.0.2 of the Advanced Light Source (ALS). Collectively, the results show that NTD-specific neutralizing antibodies all recognize a specific binding “supersite” and have adopted a convergent mechanism for inhibiting the virus.
“We did not expect that there would be a single site in the NTD that all antibodies that neutralize the virus will bind,” said McCallum. “Based on that observation, we predicted that some circulating viruses might have mutated to evade these antibodies.”
Indeed, as the researchers were writing up their manuscript and submitting it for review, variants of concern began to emerge, and before long, such mutations dominated in circulating viruses. The researchers proposed that cocktails of RBD- and NTD-binding antibodies could provide broad protection against some SARS-CoV-2 variants. Overall, the work provides evidence that NTD-targeting antibodies are a key component of immunity to SARS-CoV-2 and sheds light on the emergence of variants of concern.
M. McCallum, A. De Marco, F.A. Lempp, M.A. Tortorici, D. Pinto, A.C. Walls,M. Beltramello, A. Chen, Z. Liu, F. Zatta, S. Zepeda, J. di Iulio, J.E. Bowen, M. Montiel-Ruiz, J. Zhou, L.E. Rosen, S. Bianchi, B. Guarino, C. Silacci Fregni, R. Abdelnabi, S.-Y.C. Foo, P.W. Rothlauf, L.-M. Bloyet, F. Benigni, E. Cameroni, J. Neyts, A. Riva, G. Snell, A. Telenti, S.P.J. Whelan, H.W. Virgin, D. Corti, M.S. Pizzuto, and D. Veesler, “N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2,” Cell 184, 2332 (2021), doi:10.1016/j.cell.2021.03.028 (2021).