The use of DNA in nanotechnology applications has gained interest in recent years because it is a highly “programmable” polymer that can be precisely controlled as a building block through modification of the base-pair sequence. In addition, branched DNA motifs that are tailed by single-stranded overhangs at the end of a DNA molecule act as “sticky ends” that, like flaps of Velcro, can join two DNA molecules together, allowing the self-assembly of 2D and 3D matrices. These matrices in turn can serve as a scaffolding to organize other macromolecules, such as proteins and viruses, or as platforms to precisely control chemical processes.
In this study, the researchers designed and crystallized two related DNA scaffolds and determined their structures at ALS Beamline 8.2.2, part of the Berkeley Center for Structural Biology. They initially designed a motif that was expected to form a crystal lattice containing square cavities, with its assembly being guided by a short DNA strand at the center of the square. The resulting structure, however, instead contained a series of DNA blocks tethered together by the central strand that was originally intended to form the square. Nonetheless, the resulting structures contained a densely packed array of layers, and the 3D x-ray structures of these DNA crystals hold promise for the design of new structural motifs to create programmable 3D DNA lattices with atomic spatial resolution.
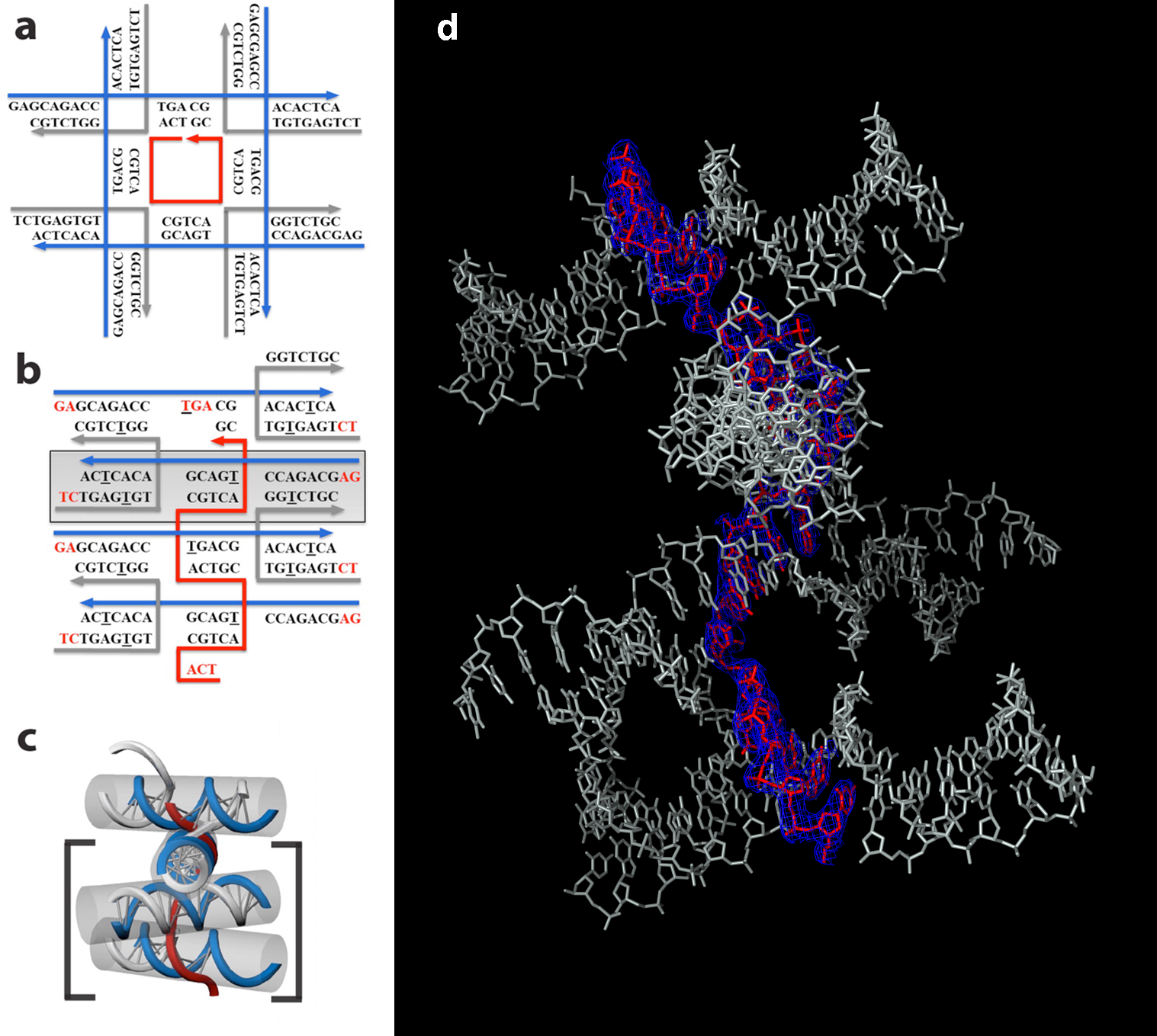
Work performed at ALS Beamline 8.2.2.
C.R. Simmons, F. Zhang, J.J. Birktoft, X. Qi, D. Han, Y. Liu, R. Sha, H.O. Abdallah, C. Hernandez, Y.P. Ohayon, N.C. Seeman, and H. Yan, “Construction and Structure Determination of a Three-Dimensional DNA Crystal,” JACS 138, 10047 (2016).