The 2024 Nobel Prize in Chemistry was awarded to David Baker, Demis Hassabis, and John M. Jumper for the development of protein structure prediction and design. At the ALS, Baker leveraged high-throughput small-angle x-ray scattering (SAXS) and protein crystallography capabilities to design novel proteins and pave a new pathway for science, technology, and the environment. Read more »
ALS Publications by David Baker
Compilation of all publications with 2024 Chemistry Nobel Laureate David Baker as co-author which used the resources of the Advanced Light Source.
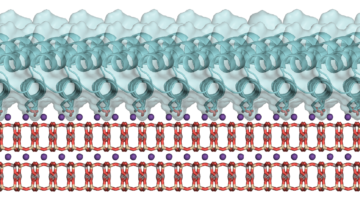
Computer-Aided Protein Design for New Biomaterials
Using a computer-based approach, researchers designed porous protein crystals that were revealed to be stable, tunable, and atomically accurate using x-ray scattering and diffraction at the ALS. The work provides a powerful new platform for biological materials engineering and opens up wide applications in biotechnology and medicine. Read more »![]()
![]()
Custom-Designed Models Reveal How Proteins Assemble on Minerals
Seashells, bone, and other hard tissues form through a little-understood process combining proteins and minerals. Researchers gained insight using a model system of proteins they designed and synthesized from scratch, characterizing how these building blocks assemble on mica. Read more »
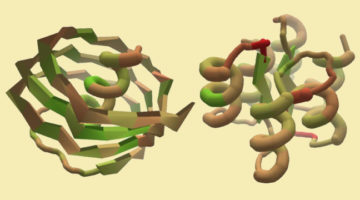
A Citizen-Science Computer Game for Protein Design
Using the computer game, “Foldit,” nonexpert citizen scientists designed new proteins whose structures, verified at the ALS, were equivalent in quality to and more structurally diverse than computer-generated designs. The work shows the potential of using crowd-based creativity in the design of new proteins for fighting illness and disease. Read more »![]()
![]()
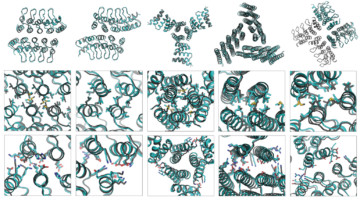
A Systematic Approach to Customizing Cyclic Proteins
Proteins consisting of identical subunits arranged symmetrically around a central axis (cyclic homo-oligomers) play key roles in many biological processes. Researchers have now developed a systematic approach to their design and demonstrated its accuracy using protein crystallography and small-angle x-ray scattering. Read more »
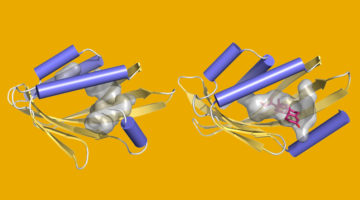
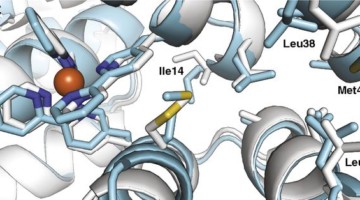
Bending the (β-Sheet) Curve to Shape Protein Cavities
Curved β sheets are basic building blocks of many protein cavities that, by serving as binding sites for other molecules, are essential to protein function. β-sheet curvature can now be controlled with atomic-level accuracy, opening the door to custom-designed sites capable of entirely new functions. Read more »![]()
![]()
NCAA Drives Formation of Designed Proteins
A noncanonical amino-acid (NCAA) complex has been found to drive the self-assembly of a computationally designed protein. Bpy-ala, which is “noncanonical” because it’s not among the 20 amino acids that occur naturally, has useful properties that could be used to generate novel photoactive proteins. Read more »
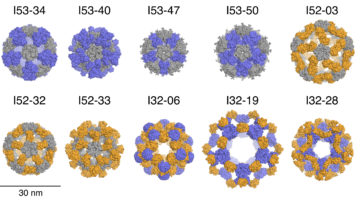
Designed Protein Nanocages Inspired by Nature
Inspired by protein molecules observed in nature, researchers have now engineered ten large, 120-subunit, two-component protein complexes. These designed nanomaterials are attractive starting points for new approaches to targeted drug delivery, vaccine design, and bioenergy. Read more »
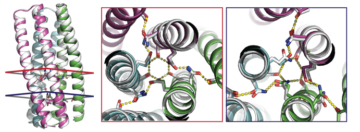
Validation of Novel Proteins Inspired by Nature
Designed proteins containing hydrogen-bonding modules have been validated by crystallography and SAXS. The ability to design synthetic molecules that combine the specificity of DNA-like binding with protein function opens up huge opportunities for the fields of synthetic biology and materials science. Read more »