Think about DNA; images of the double helix spring to mind. The double helix, however, is only one of the many fascinating structures that DNA and RNA can form. Guanine-rich DNA and RNA sequences can fold into ultra-stable quadruple-helix structures called G-quadruplexes. G-quadruplex–forming sequences are present in nearly 40% of all human genes and in many oncogenes (genes with the potential to cause cancer).
Cells must take apart or unfold double helices, G-quadruplexes, and many other structures in order to get to the genetic information stored within DNA and RNA molecules. Unfolding is accomplished by a class of proteins called helicases, which use molecules like ATP to provide the energy to either unfold, translocate, or dissociate from DNA and/or RNA.
To better understand how cells recognize and unfold G-quadruplexes, researchers used protein crystallography to visualize the unfolding of a G-quadruplex by a helicase called DHX36. Using data gathered at ALS Beamlines 5.0.1 and 5.0.2 and the Advanced Photon Source, they were able to visualize how DHX36 binds and unfolds its G-quadruplex target. DHX36 extends two recognition arms to hug the G-quadruplex, explaining the unusually high affinity and specificity DHX36 is known to display toward G-quadruplexes.
The researchers were surprised to find that, upon binding, DHX36 partially unfolds the G-quadruplex without ATP. This, coupled with another biophysical technique called single-molecule fluorescence microscopy, helped the researchers to propose an ATP-independent mechanism for DHX36-mediated G-quadruplex unfolding. The results will help researchers to better understand G-quadruplex biology and to develop therapeutic small molecules that target G-quadruplexes.
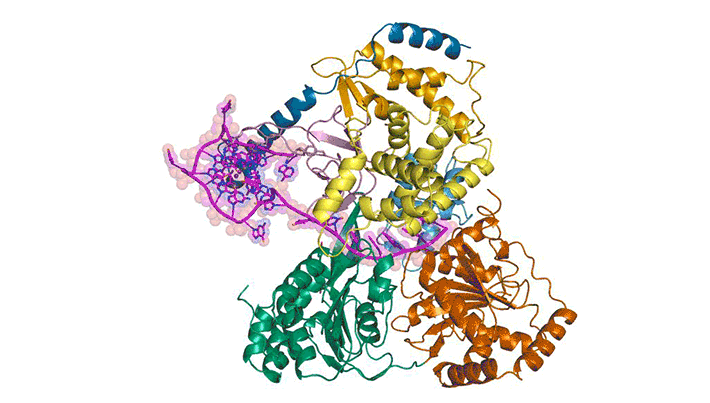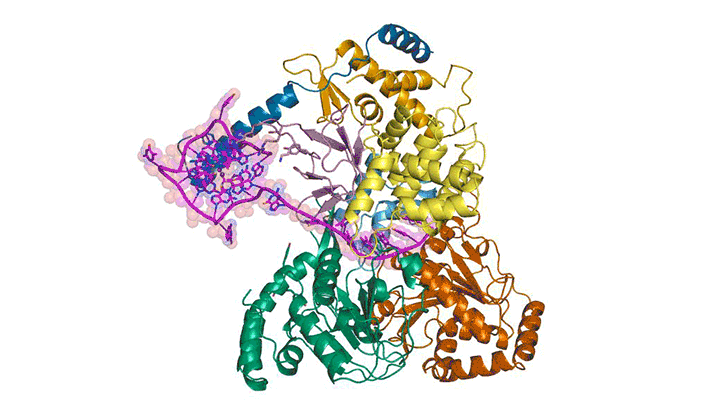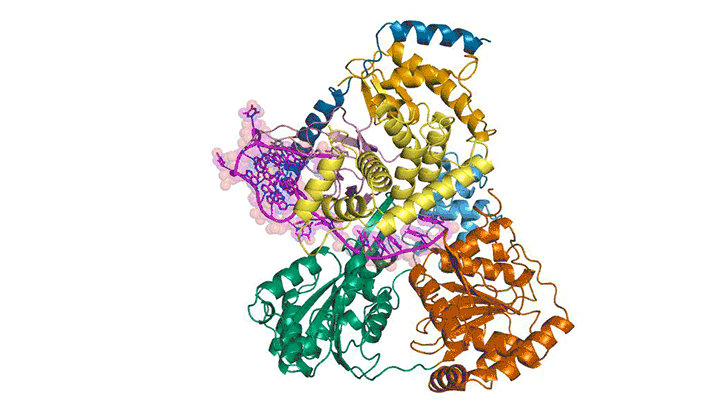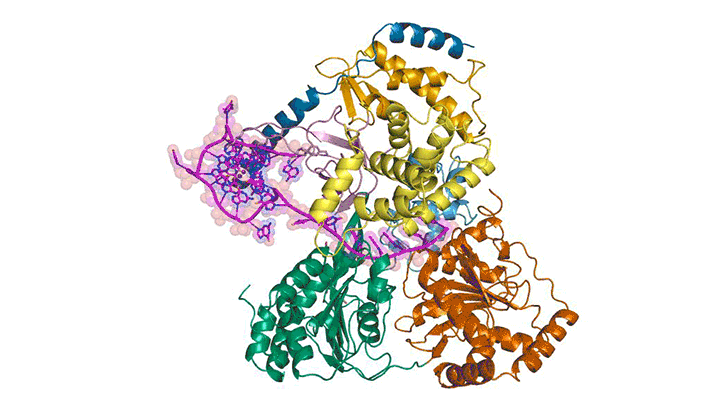
M.C. Chen, R. Tippana, N.A. Demeshkina, P. Murat, S. Balasubramanian, S. Myong, and A.R. Ferré-D’Amaré, “Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36,” Nature 558, 465 (2018), doi:10.1038/s41586-018-0209-9.