SCIENTIFIC ACHIEVEMENT
Protein structures not only revealed how SARS and MERS antibodies inhibit the viruses from attaching to host cells, they also revealed an unprecedented example of receptor mimicry that triggers the cell-invasion machinery of the SARS virus.
SIGNIFICANCE AND IMPACT
The results, obtained with the aid of protein crystallography experiments at the Advanced Light Source (ALS), inform efforts to prevent and treat these serious, often deadly, respiratory diseases.

The challenges of SARS and MERS
Severe acute respiratory syndrome (SARS) emerged in 2002, spreading across the globe and resulting in 8,000 infections and nearly 800 deaths. Middle East respiratory syndrome (MERS) emerged in 2012 and caused numerous outbreaks, with a fatality rate of 35%. Currently, there are no specific treatments or vaccines for viruses of this type (i.e., coronaviruses). Their effects range from mild cold-like symptoms to life-threatening pneumonias. In bats and other animals, coronaviruses with high genomic similarity to the human-infecting strains are constantly circulating, increasing the likelihood of future cross-species transmission. And although coronaviruses are genomically related, functionally they can be quite specialized. For example, antibodies that work against SARS won’t necessarily work against MERS, and vice versa.
A crown of spikes
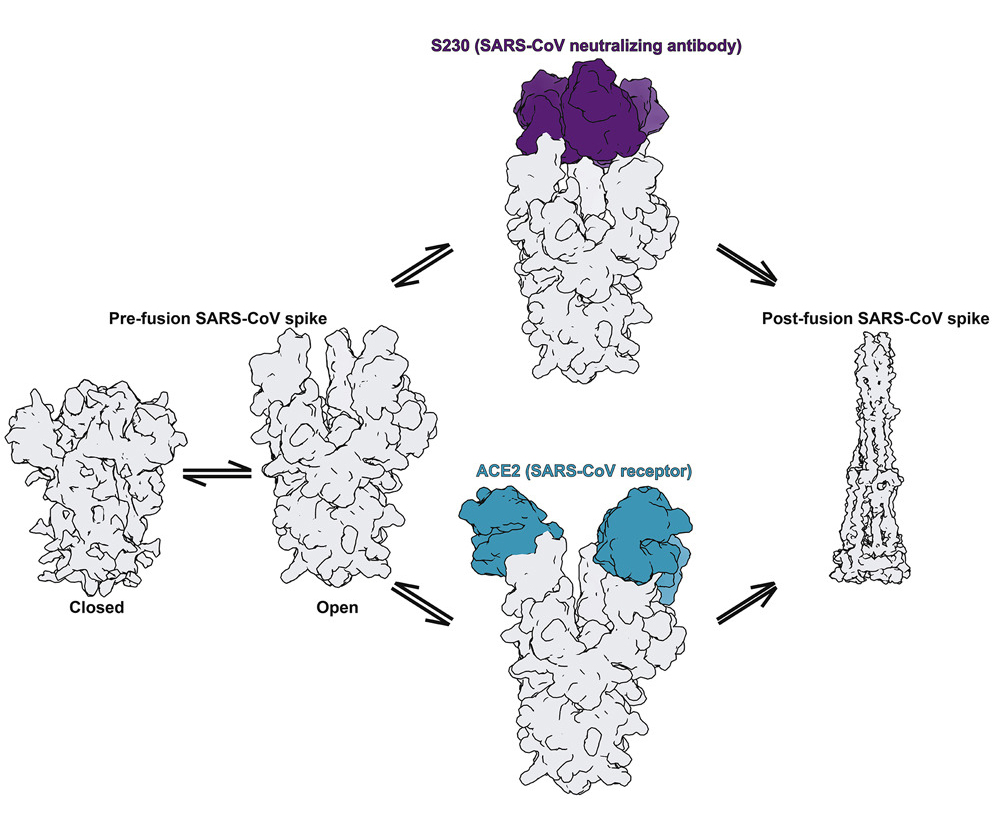
Coronaviruses are so named because they are covered with spike proteins—projections that give the appearance of a crown-like halo. These spikes facilitate fusion of the virus with a host cell, the first step in enlisting the cell’s machinery for replication. In overall architecture, spike proteins are trimers (i.e., composed of three identical subunits, or protomers) present in a metastable pre-fusion conformation at the virus surface before infection. The tip of each protomer contains a receptor-binding domain that attaches to receptors in the host-cell membrane, and each receptor-binding domain can be in an open or closed state. In addition, the spikes are “decorated” with carbohydrates (glycans) that provide camouflage from the host’s immune system. Since fusion with host cells is the first step in infection, the spikes are prime targets for both antibodies and vaccines.
A trio of techniques
To better understand how coronavirus antibodies work, a team of researchers studied spike protein structures in complex with neutralizing antibody fragments (Fabs) isolated from SARS and MERS survivors. First, they used cryo-electron microscopy (cryo-EM) and mass spectrometry to analyze the viruses’ extremely heterogeneous glycan shields. Then, to visualize how the spike structures interact with the Fabs, they used a combination of cryo-EM for the spikes (which are resistant to crystallization) and protein crystallography for the Fabs, fitting the Fab crystal structures into the cryo-EM maps. For the SARS complex in particular, which displayed a higher degree of structural variability, interpretation of the cryo-EM structure in the antibody region would not have been possible without the crystal structures. The high-resolution x-ray crystallography was performed at ALS Beamline 5.0.1, part of the Berkeley Center for Structural Biology.
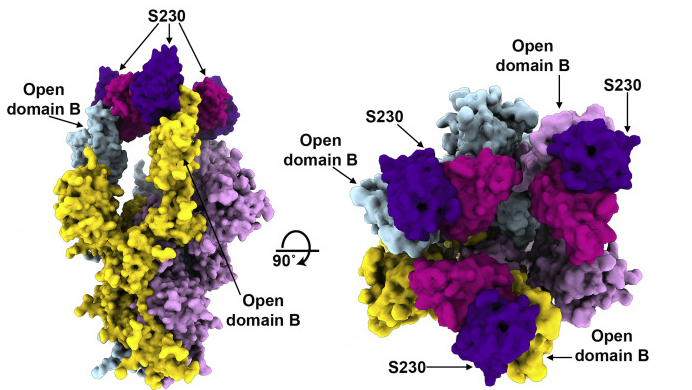
One neat antibody trick
The results showed that the MERS antibody is a potent inhibitor that interacts with spike proteins. While the same is true for the SARS antibody, the researchers also discovered an unprecedented example of functional mimcry: the SARS antibody was found to induce structural changes in the spike similar to those produced by its natural receptor (ACE2), changes known to be necessary for initiating membrane fusion. Because viruses are machines designed to work only once, triggering these changes before attachment to a host cell would effectively neutralize the virus. In general, the results provide valuable insights into how antibodies confer broad protection against coronavirus strains and inform efforts to prevent and treat these serious, often deadly, respiratory diseases.
Contact: David Veesler
Researchers: A.C. Walls, X. Xiong, Y.-J. Park, J. Snijder, J. Quispe, and D. Veesler (University of Washington); M.A. Tortorici (University of Washington and Pasteur Institute); E. Cameroni and D. Corti (Humabs Biomed, Switzerland); R. Gopal and M. Zambon (Public Health England); M. Dai (Francis Crick Institute); A. Lanzavecchia (Università della Svizzera italiana, Switzerland); and F.A. Rey (Pasteur Institute).
Funding: National Instututes of Health, Pew Charitable Trusts, Burroughs Wellcome Fund, Netherlands Organisation for Scientific Research, European Molecular Biology Organization, Zoonoses Anticipation and Preparedness Initiative, Pasteur Institute, French National Center for Scientific Research, and the University of Washington Arnold and Mabel Beckman CryoEM Center and Proteomics Resource. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences Program.
Publication: A.C. Walls, X. Xiong, Y.-J. Park, M.A. Tortorici, J. Snijder, J. Quispe, E. Cameroni, R. Gopal, M. Dai, A. Lanzavecchia, M. Zambon, F.A. Rey, D. Corti, and D. Veesler, “Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion,” Cell 176, 1026 (2019), doi:10.1016/j.cell.2018.12.028.
ALS SCIENCE HIGHLIGHT #394