Rotaviruses, the major causative agents of gastroenteritis, exhibit enormous genetic and immunological diversity. Consequently, they are classified into ten groups (A–J). Rotaviruses in group A (RVA) are the best characterized and are responsible for most gastroenteric infections worldwide, particularly in children. Rotaviruses in group B (RVB) primarily infect adults and have been associated with large epidemic outbreaks of severe gastroenteritis in China and sporadic infection elsewhere. RVB also caused recent hemorrhagic diarrheal outbreaks in piglets and foals, resulting in significant economic impact and raising the threat of potential transmission to humans.
In RVA and RVC, a domain near the tip of their spike proteins (domain VP8*) is critical for mediating the viruses’ attachment to cells. This domain exhibits a fold resembling that of versatile carbohydrate-binding proteins called galectins. Surprisingly, the VP8* of RVB (VP8*B) has a genetic sequence that differs from VP8*A and VP8*C. Little is known about the molecular mechanisms by which RVBs recognize host cells and replicate, due to the lack of a cell-culture system for RVBs and structural information about their viral proteins.
Using AlphaFold2, a deep-learning artificial-intelligence program for predicting protein structure, researchers from the Baylor College of Medicine discovered that VP8*B exhibits a novel fold distinct from the galectin-like fold. They validated this prediction by determining the structure of VP8*B to 1.3 Å resolution at Advanced Light Source Beamline 5.0.1 (part of the Berkeley Center for Structural Biology).
“Structural comparison of the predicted model and the experimental structure revealed a high degree of similarity between the models,” said Liya Hu, a researcher at the Baylor College of Medicine and first author of the study. “There were a few subtle structural differences, perhaps caused by protein–protein and protein–solvent interactions in the crystals.”
Despite the structurally distinct protein fold of VP8*B, biochemical screening showed that VP8*B performs a similar function. Thus, the study illustrates how viruses within the same family can evolve and diversify by incorporating structurally distinct modules with similar functionality. The structure of VP8*B potentially can help in the design of vaccines against the emerging group B rotaviruses.
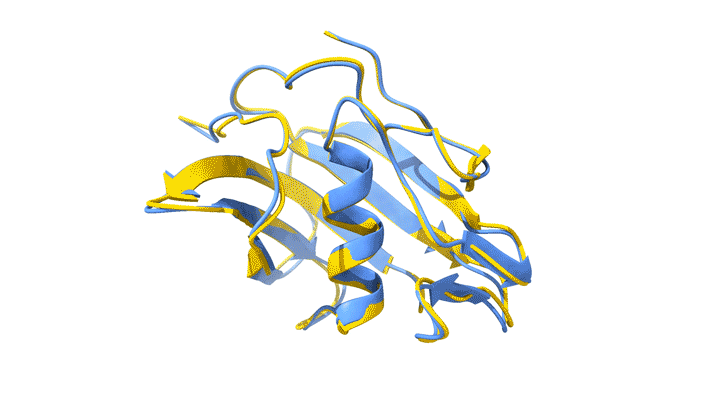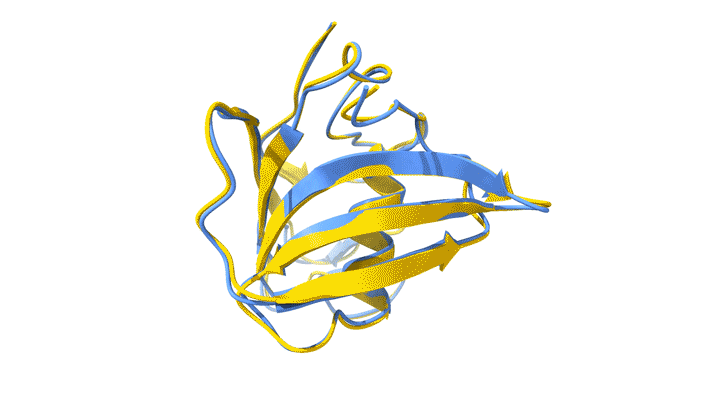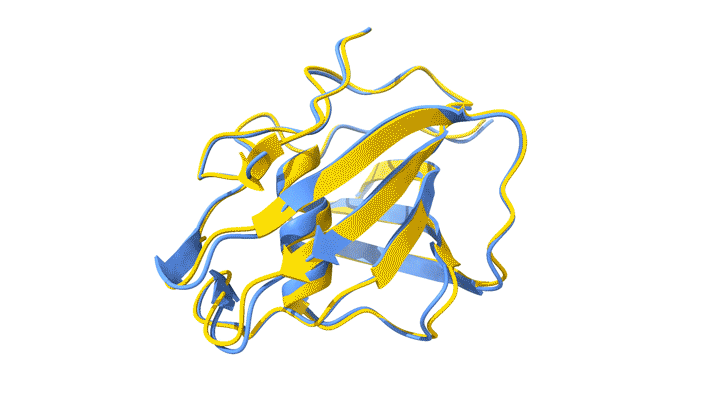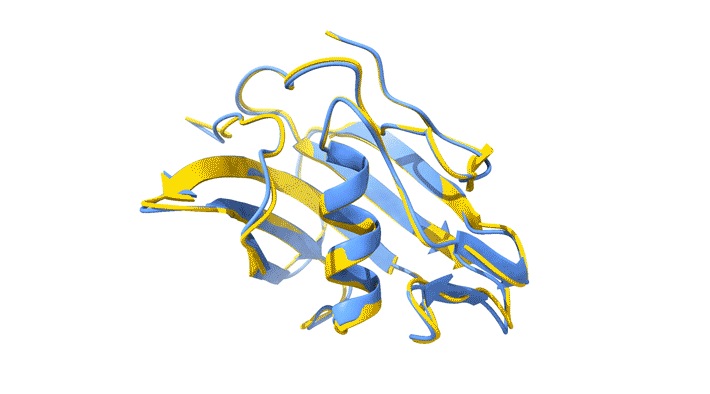
L. Hu, W. Salmen, B. Sankaran, Y. Lasanajak, D.F. Smith, S.E. Crawford, M.K. Estes, and B.V. Venkataram Prasad, “Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography,” Commun. Biol. 5, 419 (2022); doi:10.1038/s42003-022-03357-1.