SCIENTIFIC ACHIEVEMENT
Using x-ray crystallography at the Advanced Light Source (ALS) combined with biochemistry and plant genetics, researchers identified a molecular switch that triggers modifications to plant structure in response to environmental conditions.
SIGNIFICANCE AND IMPACT
A greater understanding of this adaptive process will help scientists optimize plants for efficient nutrient uptake and resistance to parasitic species.

Roots or shoots?
Anchored in soil and unable to move, plants have evolved complex mechanisms to sense and adjust to environmental conditions. For example, a plant that needs more solar energy might grow bushier shoots to collect more sunlight. On the other hand, a plant located in poor soil might develop a more extensive root system to gather scarce but essential mineral nutrients.
A key component of this adaptive process is strigolactone, a growth hormone that signals plants to divert energy into roots over shoots. When secreted from a plant’s roots, strigolactone also promotes symbiotic interactions with beneficial fungi and soil microbes. However, this plant–fungi signal can also be exploited by parasitic plants, which use it to invade and devastate crops worldwide.
Thus, to effectively minimize destructive parasitism, promote symbiotic relationships, and develop plants that are more efficient at nutrient uptake, scientists need a molecular-level understanding of the strigolactone-based signaling mechanism.
An enzymatic toggle switch
Strigolactone is known to act in concert with several enzymes (molecules that join, stabilize, or break up other molecules). In 2018, research groups from the University of Washington and the University of California, Davis, broke new ground on this topic by discovering how one key enzyme rearranges itself to interact with strigolactone and other associated compounds.
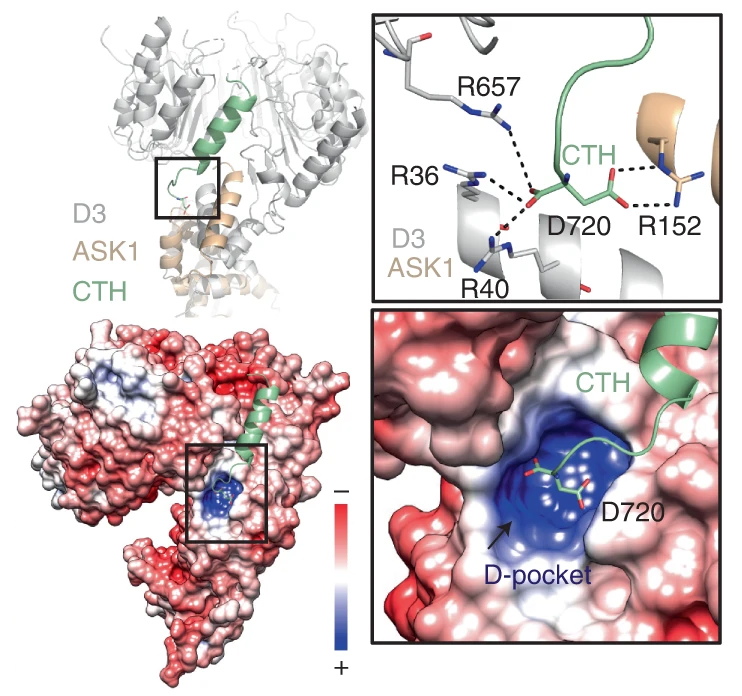
The enzyme, called MAX2 in the plant model Arabidopsis thaliana, was found to toggle between locked and unlocked conformations. When unlocked, MAX2 forms a complex with strigolactone and a sensor molecule, D14. The MAX2–strigolactone–D14 complex facilitates the destruction of D53 proteins that suppress the expression of genes promoting root development. With D53 out of the picture, a plant will more aggressively develop its root system.
In this work, the researchers from UC Davis systemically mutated the MAX2 enzyme and created a version fixed in its unlocked form. Once they verified that the purified protein was functional and in a distinct conformational state, they set up extensive crystallization trials to determine its 3D molecular structure.

Switch mechanism and trigger
The x-ray crystallography was conducted at ALS Beamlines 8.2.1 and 8.2.2, part of the Berkeley Center for Structural Biology (BCSB). The state-of-the-art remote capabilities enabled the UC Davis researchers to quickly process multiple crystal datasets to determine the crystal structure of the MAX2 enzyme.
The data allowed identification of the mechanism behind the enzyme’s activity in strigolactone signaling. The unlocked conformation involves shifts in a C-terminal helix, which enables D53 repressor proteins to be tagged for destruction. To release the D53 for disposal, the enzyme must relock. This in turn causes the D14 strigolactone sensor to also be tagged for destruction, providing feedback for regulating the signaling cascade.
Finally, the study also revealed the key to the MAX2 lock. An organic acid metabolite, citrate, can directly trigger the conformational shift in the C-terminal helix. The citrate is likely trapped in a highly charged pocket that’s exposed when the enzyme is unlocked. This is the first time that a primary metabolite has been shown to act as a direct regulator of this type of enzyme and points to interesting avenues for future study.
Contact: Nitzan Shabek
Researchers: L. Tal, M. Palayam, M. Ron, A. Young, A. Britt, and N. Shabek (University of California, Davis).
Funding: National Science Foundation, National Institutes of Health, and United States–Israel Binational Agricultural Research and Development Fund. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences program.
Publication: L. Tal, M. Palayam, M. Ron, A. Young, A. Britt, and N. Shabek, “A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling,” Nat. Plants 8, 561 (2022), doi:10.1038/s41477-022-01145-7.
Adapted from the UC Davis blog post, “Unlocked Enzyme Structure Shows How Strigolactone Hormone Controls Plant Growth,” and the Berkeley Lab Biosciences Area highlight, “Enzyme Structure Reveals Key Details in Strigolactone Signaling.”
ALS SCIENCE HIGHLIGHT #468