SCIENTIFIC ACHIEVEMENT
Protein-structure studies performed in part at the Advanced Light Source (ALS) helped researchers discover that the protein assemblies in a key carbon-cycling enzyme can rearrange with surprising ease.
SIGNIFICANCE AND IMPACT
The findings raise the prospect of genetically tuning the enzyme in agricultural plant species to produce more productive and resource-efficient crops.
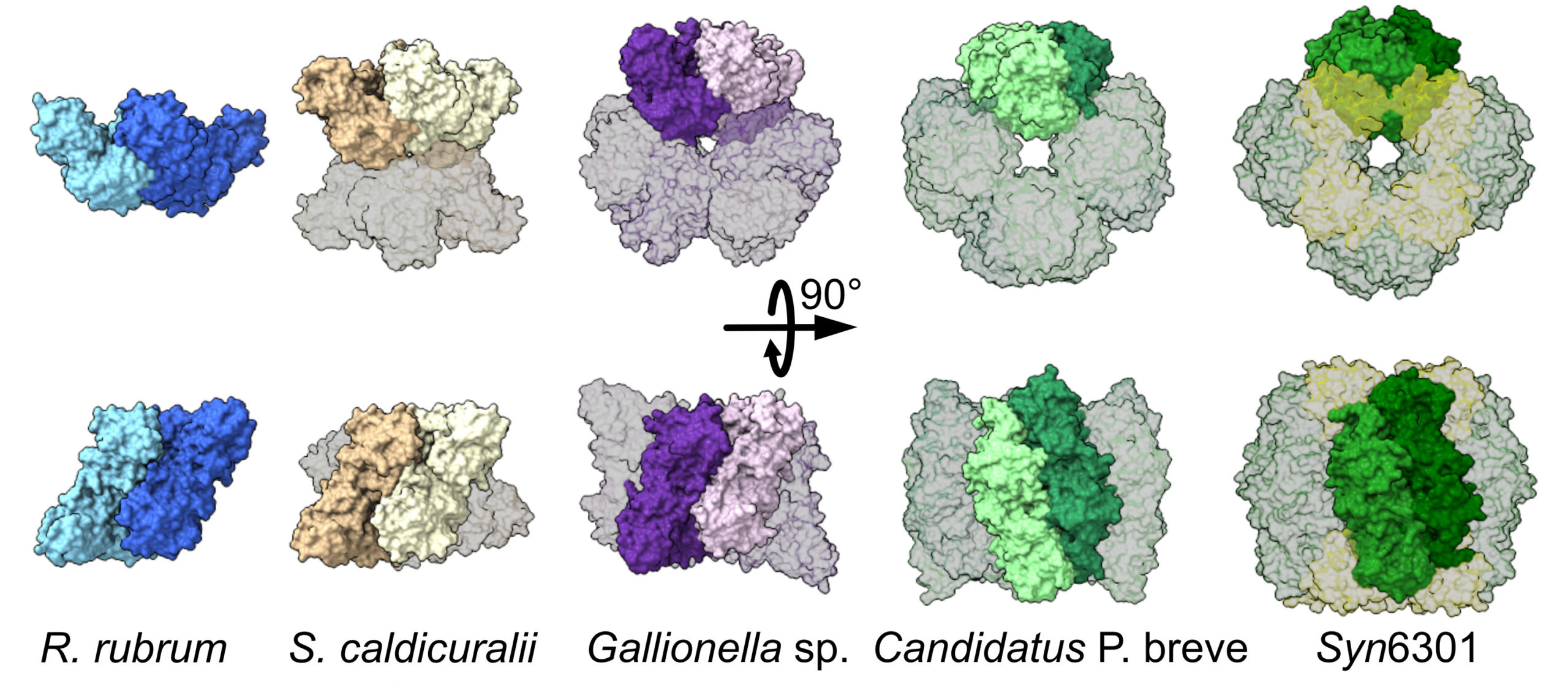
Structural plasticity in rubisco
One of the key steps to understanding biology is discovering how a protein does its job. This requires knowledge of protein structures, from the atomic level to that of multiprotein assemblies. In a study of rubisco—an enzyme that enables photosynthetic organisms to convert CO2 in the atmosphere into biomass—researchers showed how evolution can lead to a surprising diversity of molecular assemblies that all accomplish the same task.
In this work, the researchers studied a type of rubisco (form II) found in bacteria and a subset of photosynthetic microbes. At the ALS, the study combined the atomic resolution of traditional crystallography with small-angle x-ray scattering (SAXS), a high-throughput process that can take snapshots of proteins in their native form in liquid mixtures.
A diversity-driven approach
Historically, if scientists solved a protein structure crystallographically and determined that it was dimeric (composed of two units), they might assume that similar proteins would also be dimeric. But small sample sizes and sampling bias—unavoidable given the difficulty of crystallizing proteins—obscures reality. A broader approach is needed to capture the structural diversity found within protein families as well as changes over time.
Previous work has shown that the type of rubisco found in plants (form I) has an octameric core, whereas form II was believed to exist mostly as a dimer with a few rare examples of hexamers. Here, the researchers used a number of analytical approaches—structural, genomic, biochemical, and computational—to investigate the evolutionary trajectory of form II rubisco.
Synchrotron-based experiments
At ALS Beamline 12.3.1, the researchers analyzed rubisco from a diverse range of microbe species using size-exclusion chromatography coupled with small-angle x-ray scattering and multiangle light scattering (SEC-SAXS-MALS). This allowed structural differentiation via x-ray scattering profiles and measured molecular weights. The results revealed mostly hexameric rubisco, with a handful in the dimeric state.
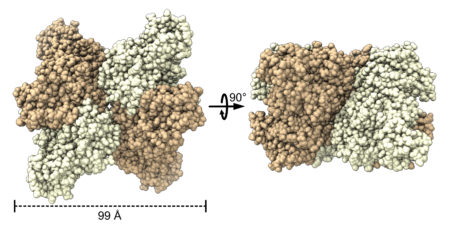
Notably, the data revealed the presence of a novel tetrameric form of rubisco. Based on earlier studies that suggested tetrameric rubisco might be found in the bacterium S. caldicuralii, the researchers performed crystallographic studies at ALS Beamline 5.0.2 to confirm this. Subsequent crystallography at the National Synchrotron Light Source II (NSLS‑II) supported investigations into how rubisco’s activity is affected by mutations that facilitate its transformation from dimer to hexamer.
Where it started, where it’s going
With the structural data, the team was able to perform ancestral sequence reconstruction—a computer-based method that can estimate what ancestral proteins looked like based on the sequence and appearance of modern proteins. The reconstruction suggests that the gene for form II rubisco has changed over its evolutionary history to produce proteins with a range of structures that transform into new shapes or revert back to older structures quite easily, facilitating evolution and innovation.
Moreover, the mutational experiments unexpectedly produced a form of rubisco that is better at utilizing its target molecule, CO2. This raises the prospect of genetically tuning the rubisco in agricultural plant species to increase the protein’s affinity for CO2, in order to produce more productive and resource-efficient crops. The work highlights the power of combining genomic and structural information to study one of the most important problems in biology.

Contact: Patrick Shih
Researchers: A.K. Liu (UC Berkeley, UC Davis, and Berkeley Lab); J.H. Pereira (Joint BioEnergy Institute and Berkeley Lab); A.J. Kehl, S.K.S. Chu, and J.B. Siegel (UC Davis); D.J. Rosenberg (UC Berkeley and Berkeley Lab); D.J. Orr (Lancaster Environment Centre, UK); D.M. Banda and M. Hammel (Berkeley Lab); P.D. Adams and P.M. Shih (Joint BioEnergy Institute, UC Berkeley, and Berkeley Lab).
Funding: Branco Weiss fellowship (ETH Zürich); David and Lucile Packard Foundation; U.S. Department of Energy (DOE), Office of Science, Biological and Environmental Research program; National Institutes of Health; and Biotechnology and Biological Sciences Research Council (UK). Operation of the ALS and NSLS-II is supported by DOE, Office of Science, Basic Energy Sciences program.
Publication: A.K. Liu, J.H. Pereira, A.J. Kehl, D.J. Rosenberg, D.J. Orr, S.K.S. Chu, D.M. Banda, M. Hammel, P.D. Adams, J.B. Siegel, and P.M. Shih, “Structural plasticity enables evolution and innovation of RuBisCO assemblies,” Sci. Adv. 8, eadc9440 (2022), doi:10.1126/sciadv.adc9440.
Adapted from the Berkeley Lab news release, “Protein Structures Aren’t Set in Stone.”
ALS SCIENCE HIGHLIGHT #474