The Zika virus (ZIKV) is a mosquito-borne pathogen recently linked to certain birth defects in infants in South and Central America and the United States. Protein crystallography studies performed at the ALS have now resolved the structure of a key ZIKV protein (NS5) to 3.0 Å. The high-resolution structure has enabled researchers to make detailed comparisons to similar proteins from related viruses and gain insight into how their differences might affect viral replication and immune response. The availability of this structure is an important step toward the targeted, structure-based development (and repurposing) of drugs capable of disrupting viral functions and halting the spread of the disease.

ZIKV belongs in the genus Flavivirus, which derives its name from the virus that causes yellow fever (flavus means “yellow” in Latin). Other members of the Flaviviridae family include the human pathogens that cause Japanese encephalitis (JEV) and dengue fever (DENV). Expression of flavivirus genomes results in the production of three proteins that are part of the virus itself (structural proteins) and seven proteins that are not part of the virus, but could perform some viral function in the infected cell (nonstructural proteins).
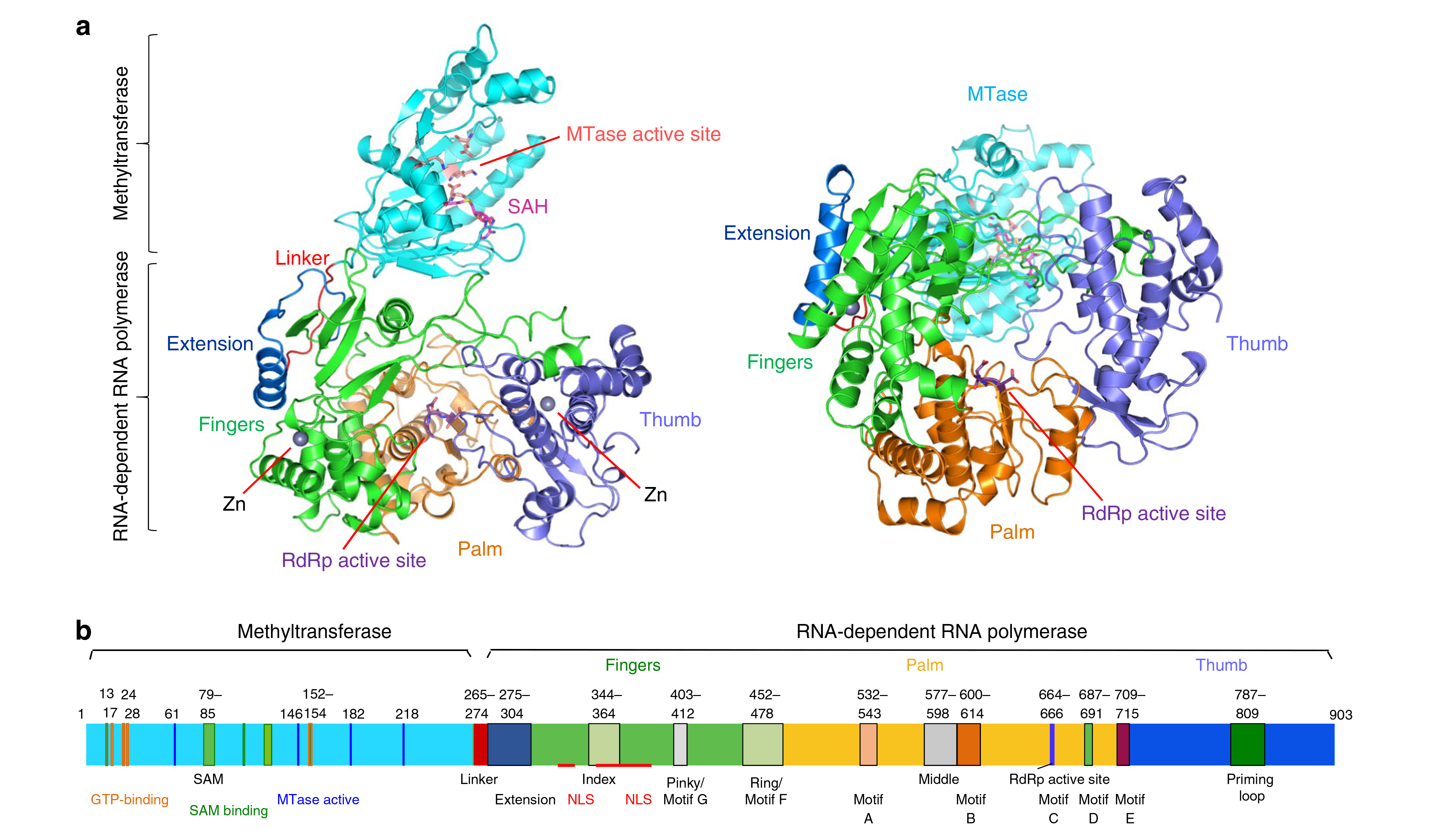
In ZIKV, nonstructural protein 5 (NS5) includes two domains: one that facilitates the translation of the viral genome and helps the virus to evade the host’s immune response (the methyltransferase domain, MT) and another that helps start the genetic replication process (the RNA-dependent RNA polymerase domain, RdRp). Because of these crucial functions, NS5 in ZIKV is a potent drug target whose analogues in Japanese encephalitis and dengue fever have been used to develop therapies that work at the molecular level.
To study the ZIKV NS5 structure in detail, the researchers first produced and crystallized small quantities of the full-length NS5 protein using chemically synthesized DNA that encoded the ZIKV strain originally isolated from Uganda, Africa, in 1947 (strain MR766). X-ray crystallography at ALS Beamline 5.0.2, part of the Berkeley Center for Structural Biology, was then used to solve the atomic structure. The powerful beam and the sensitive detector made it possible to obtain good data despite less-than-optimal crystal quality. In addition, access to the Collaborative Crystallography program at the ALS—a fast, reliable, and transparent mail-in crystallographic service funded by the National Institutes of Health—resulted in rapid turnaround for this time-sensitive project.
The solved structure revealed remarkable similarities to the equivalent structures of other viruses from the Flaviviridae family, the active sites in particular. The results suggest that inhibitors of viral MT activity and/or RNA synthesis in related viruses can be developed to inhibit ZIKV replication as well. In fact, molecules that can inhibit RNA synthesis in dengue virus, West Nile virus, and yellow fever virus have recently been shown to affect RNA synthesis by ZIKV NS5. Solving additional structures of the ZIKV NS5 complexed to such molecules could aid in the development of inhibitors with higher specificity and potency.
While the Zika virus was initially detected in 1947 in Uganda, serious illness associated with Zika virus infection was not recorded until 2007. At present, the basis for this escalation in virulence with the more recent Zika virus outbreak remains to be established. However, comparison of recombinant NS5 proteins from Africa and from Brazil revealed similar levels of RNA synthesis. In addition, the residues of the Brazilian ZIKV that differ from those of the MR766 virus from Africa are mostly on the surface of the NS5 protein and are less likely to affect the mechanism of RNA-dependent RNA synthesis. The changes, however, could impact interactions with other ZIKV proteins or with cellular proteins. Thus, while the researchers have gathered a great deal of information about how to target this protein, there are still puzzles remaining. Further studies will be invaluable for therapeutics discovery.
Contacts: Pingwei Li and C. Cheng Kao
Research conducted by: B. Zhao, F. Du, and P. Li (Texas A&M University); G. Yi, Y.-C. Chuang, R.C. Vaughan, and C.C. Kao (Indiana University); and B. Sankaran (Berkeley Lab).
Research funding: Johnson Center for Innovation and Translational Research, National Institutes of Health, and Howard Hughes Medical Institute. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences Program.
Publication about this research: B. Zhao, G. Yi, F. Du, Y.-C. Chuang, R.C. Vaughan, B. Sankaran, C.C. Kao, and P. Li, “Structure and function of the Zika virus full-length NS5 protein,” Nat. Commun. 8, 14762 (2017). doi:10.1038/ncomms14762
Adapted from the Berkeley Lab news release, “Researchers Gain Insight into Protein Critical to Zika Virus Reproduction.”
ALS SCIENCE HIGHLIGHT #357