Over the course of billions of years, nature has evolved particular molecular structures that form the basis of life, such as those found in nucleic acids and proteins. Using the natural form as a springboard, University of Washington researchers have designed protein homo-oligomers, or identical interacting subunits, that can contain interchangeable hydrogen-bonding modules for building different structures or functions.
The research group had about 100 molecules that had been expressed from synthetic genes in bacteria, all computationally designed from scratch (“de novo”) using Rosetta modeling software and a new method, HBNet. Of these, ten representative molecules were selected for x-ray crystallographic structure determination at ALS Beamlines 8.2.1 and 8.2.2 of the Berkeley Center for Structural Biology (BCSB). Those designs not chosen for x-ray crystallography were studied using small-angle x-ray scattering (SAXS) at the Structurally Integrated Biology for Life Sciences (SIBYLS) Beamline 12.3.1.
The x-ray crystallography showed that the structures and hydrogen-bond networks within the proteins were in good agreement with the predicted models. SAXS provided further validation of the structures, as well as fundamental information about the degree of agreement between the proteins and models for the remaining predicted formations. The ability to design synthetic molecules that combine the specificity of DNA-like binding with protein function opens up huge opportunities for the fields of synthetic biology and materials science.
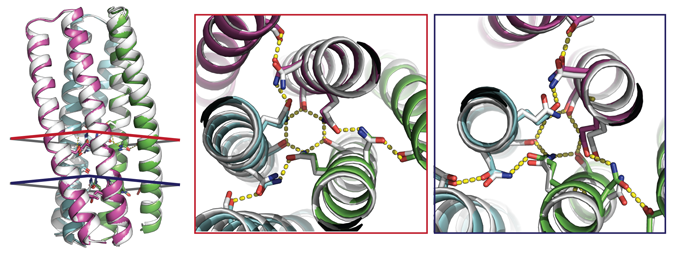
Work performed at ALS Beamlines 8.2.1 and 8.2.2 (Berkeley Center for Structural Biology) and 12.3.1 (Structurally Integrated Biology for Life Sciences).
S.E. Boyken, Z. Chen, B. Groves, R.A. Langan, G. Oberdorfer, A. Ford, J.M. Gilmore, C. Xu, F. DiMaio, J.H. Pereira, B. Sankaran, G. Seelig, P.H. Zwart, and D. Baker, “De novo design of protein homo-oligomers with modular hydrogen-bond network–mediated specificity,” Science 352, 680 (2016).