SCIENTIFIC ACHIEVEMENT
Researchers discovered a bacterial enzyme, structurally characterized at the Advanced Light Source (ALS), that synthesizes a biopolymer whose repeating units are linked together in a way that had not been previously observed.
SIGNIFICANCE AND IMPACT
The new polymer is biodegradable and may be biocompatible, with potential for applications ranging from medical therapeutics to eco-friendly plastic alternatives.
A serendipitous search
Databases of DNA sequences from a variety of organisms are treasure troves of data, and they’re growing faster than scientists can sift through them. As part of a project to design high-throughput methods to screen and characterize genomic data, researchers from the University of British Columbia focused on discovering enzymes capable of breaking down chitin, an abundant biopolymer found in insect and crustacean exoskeletons that might be economically converted into useful biochemicals.
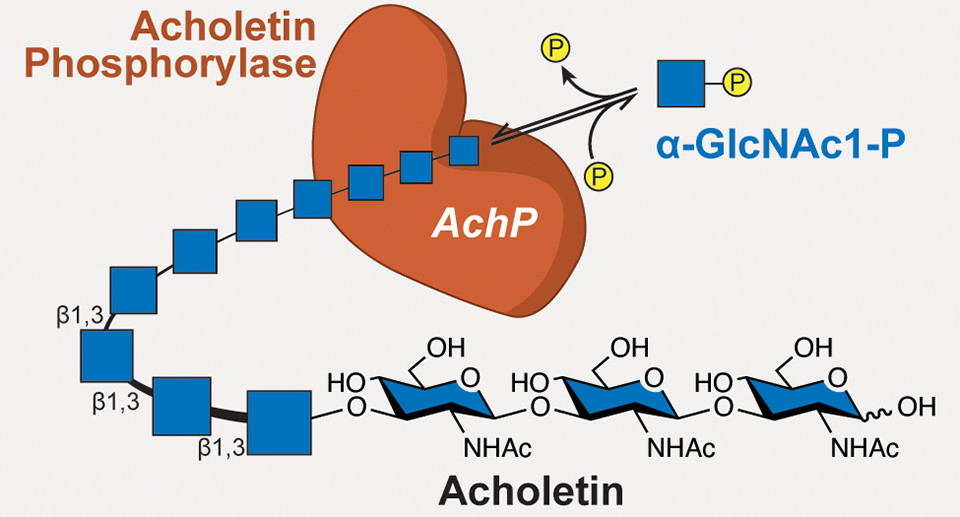
What the researchers discovered, however, was an enzyme, sourced from the bacterium Acholeplasma laidlawii, that produces a biopolymer with the same building blocks as chitin, but linked in a different way. The new polymer, dubbed acholetin (combining Acholeplasma and chitin), has the potential to be a new type of biocompatible and biodegradable polymer that could be used in a wide variety of ways, from drug delivery systems that won’t trigger immune responses to alternatives to synthetic polymers (plastics) derived from fossil fuels.
Finding the missing link
About 200 genes encoding enzymes of interest were synthesized for the researchers by the Joint Genome Institute at Berkeley Lab. The resulting enzymes were biochemically screened to find ones that act on GlcNAc, chitin’s basic carbohydrate building block. One enzyme was found to form polymers out of GlcNAc precursors, but it couldn’t perform the reverse reaction, cleaving chitin strands into GlcNAc subunits. This was curious because, in the enzymes being screened, activities are usually reversible, with the reaction direction driven by the relative concentrations of precursor and product.
The researchers thought that the difference between chitin and the new polymer, acholetin, might lie in the type of bonds linking the GlcNAc subunits. GlcNAc is a cyclic carbohydrate, with six carbons in a ring, numbered 1 to 6. In chitin, the linkage is via carbons 1 and 4 (a β-1,4 glycosidic linkage). Acholetin, they hypothesized, was linked through carbons 1 and 3, the only chemically possible GlcNAc linkage not yet reported in the literature. Nuclear magnetic resonance spectrometry confirmed that the enzyme (acholetin phosphorylase, or AchP) generates β-1,3 glycosidic linkages.
Analysis of enzyme structure
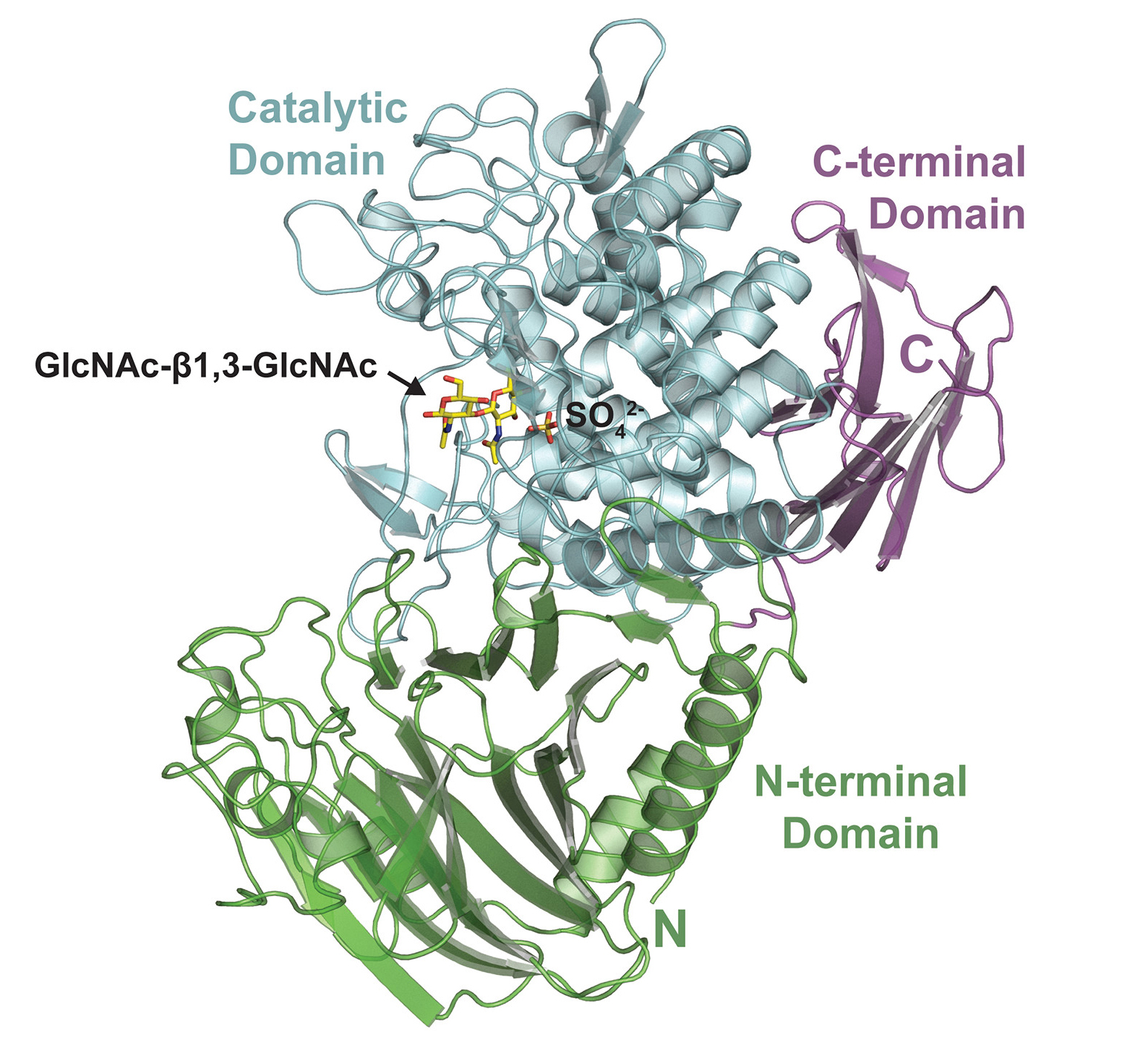
To gain insight into how AchP works, structures of the enzyme together with GlcNAc substrates (with either the enzyme or the substrate in an inactive form) were visualized using x-ray crystallography at ALS Beamlines 5.0.2 and 8.2.2. The results revealed that, in its natural form, AchP likely acts as a dimer, with parts of each monomer contributing to the formation of an active site that binds GlcNAc-β-1,3-GlcNAc. The structures also showed the GlcNAc in a transitional conformation, which fits well with how the catalytic mechanism of these enzymes is understood to work. In general, the structural details helped validate the biochemical results and deepened understanding of how AchP makes acholetin.
In principle, the enzymatic production of acholetin is scalable, but in practice it’s limited by the cost of producing the required starting ingredient. Ironically, the discovery of an enzyme that breaks down chitin—the researchers’ original goal—would make production on an industrial scale economical. Thus, in addition to analyzing acholetin’s material properties, the researchers plan to continue the search for that elusive chitin-breaking enzyme.
Contact: Spencer Macdonald
Researchers: S.S. Macdonald, F. Liu, G. Tegl, J.F. Wardman, and S.G. Withers (University of British Columbia); J.H. Pereira and A. DeGiovanni (Joint BioEnergy Institute and Berkeley Lab); S. Deutsch and Y. Yoshikuni (Joint Genome Institute, Berkeley Lab); and P.D. Adams (Joint BioEnergy Institute, Berkeley Lab, and UC Berkeley).
Funding: Natural Sciences and Engineering Research Council of Canada; Canadian Institutes for Health Research; Howard Hughes Medical Institute; National Institutes of Health; and U.S. Department of Energy (DOE), Office of Science, Biological and Environmental Research program. Operation of the ALS is supported by DOE Office of Science, Basic Energy Sciences program.
Publication: S.S. Macdonald, J.H. Pereira, F. Liu, G. Tegl, A. DeGiovanni, J.F. Wardman, S. Deutsch, Y. Yoshikuni, P.D. Adams, and S.G. Withers, “A Synthetic Gene Library Yields a Previously Unknown Glycoside Phosphorylase That Degrades and Assembles Poly-β-1,3-GlcNAc, Completing the Suite of β-Linked GlcNAc Polysaccharides,” ACS Cent. Sci. 8, 430 (2022), doi:10.1021/acscentsci.1c01570.
ALS SCIENCE HIGHLIGHT #464
See also the Berkeley Lab Science Snapshot, “Newly Discovered Bacterial Enzyme Produces Useful Biopolymer.”