Bacterial microcompartments (BMCs) are hollow protein shells that encapsulate enzymes involved in bacterial metabolism. They serve to co-localize the enzymes and their reactants for greater efficiency, as well as to sequester reaction products from the rest of the cell. Despite the availability of structural information on individual shell components, the principles governing how the pieces fit together have remained elusive. Researchers have now performed protein crystallography studies at the ALS and at Stanford Synchrotron Radiation Lightsource (SSRL) of a fully assembled BMC as well as its separate building blocks. The resulting atomic-resolution views reveal the basic principles of shell construction and provide important information for fighting pathogens and for bioenergy or biotechnology applications.

The first BMCs to be discovered were used by some bacteria to metabolize (or “fix”) carbon dioxide, similar to the way plants combine carbon dioxide with sunlight (via photosynthesis) to produce food or fuel. Understanding how these microcompartments are assembled, as well as how they let some compounds pass through while impeding others, could contribute to research in enhancing carbon fixation and, more broadly, bioenergy. BMCs also help many types of pathogenic bacteria metabolize compounds that are not available to normal, non-pathogenic microbes, giving the pathogens a competitive advantage.
The contents of a given BMC determine its specific function, but the overall architecture of BMCs are fundamentally the same. The microcompartment shell provides a selectively permeable barrier which separates the reactions in its interior from the rest of the cell. This enables higher efficiency in multi-step reactions, prevents undesired interference, and confines toxic compounds that may be generated by the encapsulated reactions.
Although BMCs are functionally similar to the membranes that surround organelles in plant and animal cells, these bacterial analogues are structurally different: they are polyhedral shells made of proteins rather than sacs made of lipids. To allow selected molecules to pass through, lipid-based membranes are dotted with proteins that create pores. BMC shells, on the other hand, are already made of proteins. Thus, these constituent proteins not only play a structural role, they are also responsible for selectively channeling molecules in and out.
Earlier studies had revealed the structures of some individual components that make up the BMC shell, but imaging the whole thing was challenging because of its large mass of about 6.5 megadaltons, roughly equivalent to the mass of 6.5 million hydrogen atoms. This size of protein compartment can contain up to 300 average-sized proteins. Researchers crystallized the BMCs from Haliangium ochraceum (ocean-dwelling slime bacteria) and collected x-ray diffraction data at ALS Beamline 5.0.2 and SSRL Beamline 12.2.
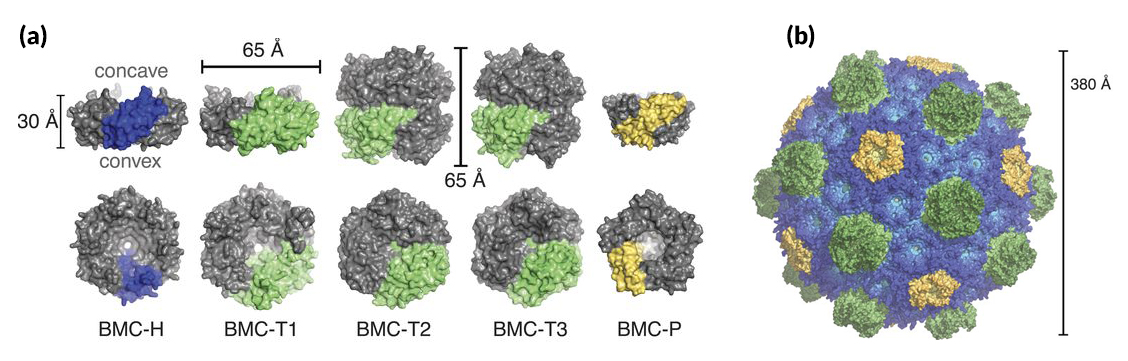
The complete shell structure shows how three different kinds of shapes—pentagons, hexagons, and double-stacked hexagons—self-assemble into a 20-sided polyhedron, with the pentagons at the vertices and the hexagons forming the facets. It answers the fundamental questions of whether the shell is single or double layered (single), how the stacked hexagons are accommodated (the “extra” hexagon protrudes to the outside), and how the individual components are oriented (concave side out). The angles between the various components were measured, and their side-chain interactions were catalogued. The intact shell was shown to be tightly packed, with the only way in or out being through pores formed at the centers of the hexagons. The pores in the double-stacked hexagons could be open in one layer open and closed in the other. Other details influencing the curvature and final size of the compartment were elucidated, resulting in scaling principles for constructing a range of shell sizes.
Given the high degree of similarity among shell proteins of all BMCs, these principles apply to functionally diverse microcompartments across the bacterial kingdom and can inform the design and engineering of shells with new functionalities.
Contact: Cheryl Kerfeld
Research conducted by: M. Sutter and C.A. Kerfeld (Michigan State University and Berkeley Lab), B. Greber (Berkeley Lab and UC Berkeley), and C. Aussignargues (Michigan State University).
Research funding: National Institutes of Health, Swiss National Science Foundation, and the U.S. Department of Energy, Office of Science, Basic Energy Sciences Program (DOE BES). Operation of the ALS is supported by DOE BES.
Publication about this research: M. Sutter, B. Greber, C. Aussignargues, and C.A. Kerfeld, “Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell,” Science 356, 1293 (2017). doi:10.1126/science.aan3289
Adapted from the Berkeley Lab news release, “Study Sheds Light on How Bacterial Organelles Assemble.”
ALS SCIENCE HIGHLIGHT #358