SCIENTIFIC ACHIEVEMENT
Multiscale, multimodal visualization techniques at the Advanced Light Source (ALS) enabled researchers to clarify how proteins remodel bacterial DNA in response to stressful environments.
SIGNIFICANCE AND IMPACT
The discovery could lead to new strategies for controlling microbial behavior and, eventually, new ways to fight bacterial infections.
Bacterial rapid response
When bacteria are put in different environments, such as one that is more acidic or anaerobic, their genes start to adapt remarkably quickly. They’re able to do so because the proteins making up their chromosomes can pack and unpack rapidly, regulating the expression of genes that could help them respond to the changing environment. Now, researchers have visualized this process at the molecular level using advanced imaging techniques, an achievement that could eventually enable scientists to develop strategies to control microbial behavior.
DNA-packing proteins
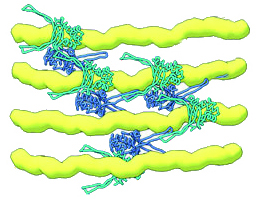
In bacteria, the proteins responsible for DNA packing are called HU proteins. They are dimers—made up of two identical or nearly identical subunits that, when joined, form a bilaterally symmetrical “body” with two “arms.” To perform their packing function, these dimers “hug” the DNA, with arms on either side of a strand. They link up with each other (multimerize) to form arrays that hold the DNA strands parallel to each other. Changes that loosen or tighten this DNA bundling make the genetic information in the DNA more or less accessible to the enzymes responsible for gene expression.
In earlier work, the researchers used the ALS’s complementary visualization capabilities to show how HU proteins can trigger pathogenicity in E. coli. In this work, the researchers focused on how external environmental factors, such as acidity or salinity, affect HU function.
Visualization in three ways
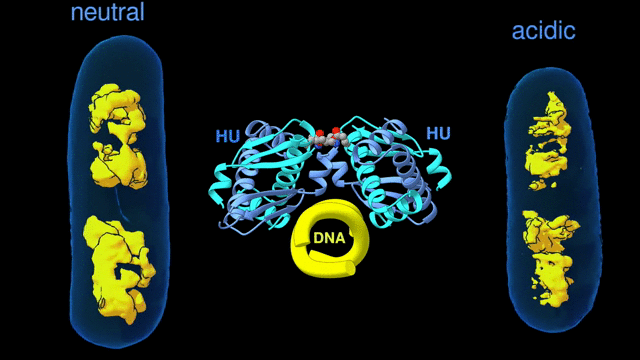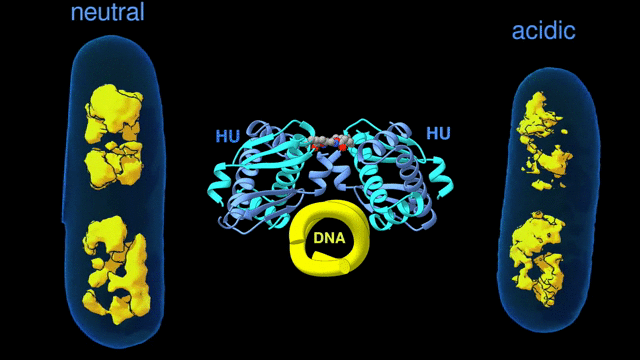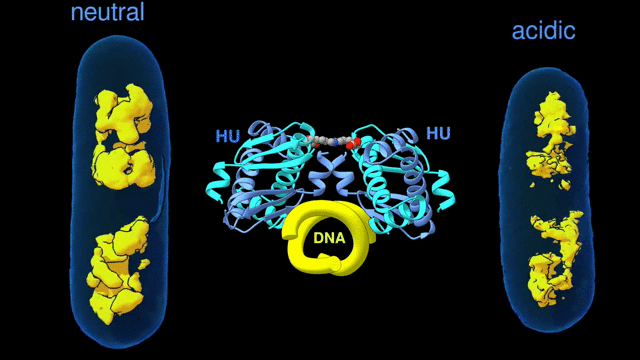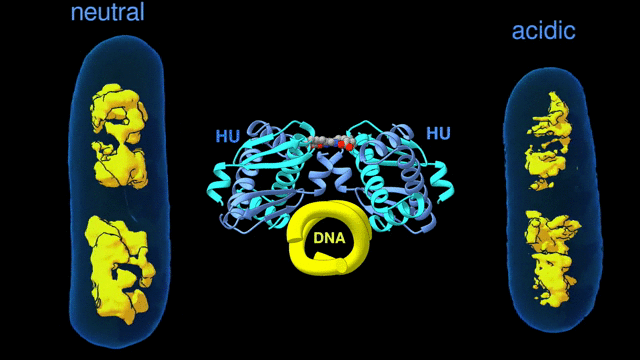
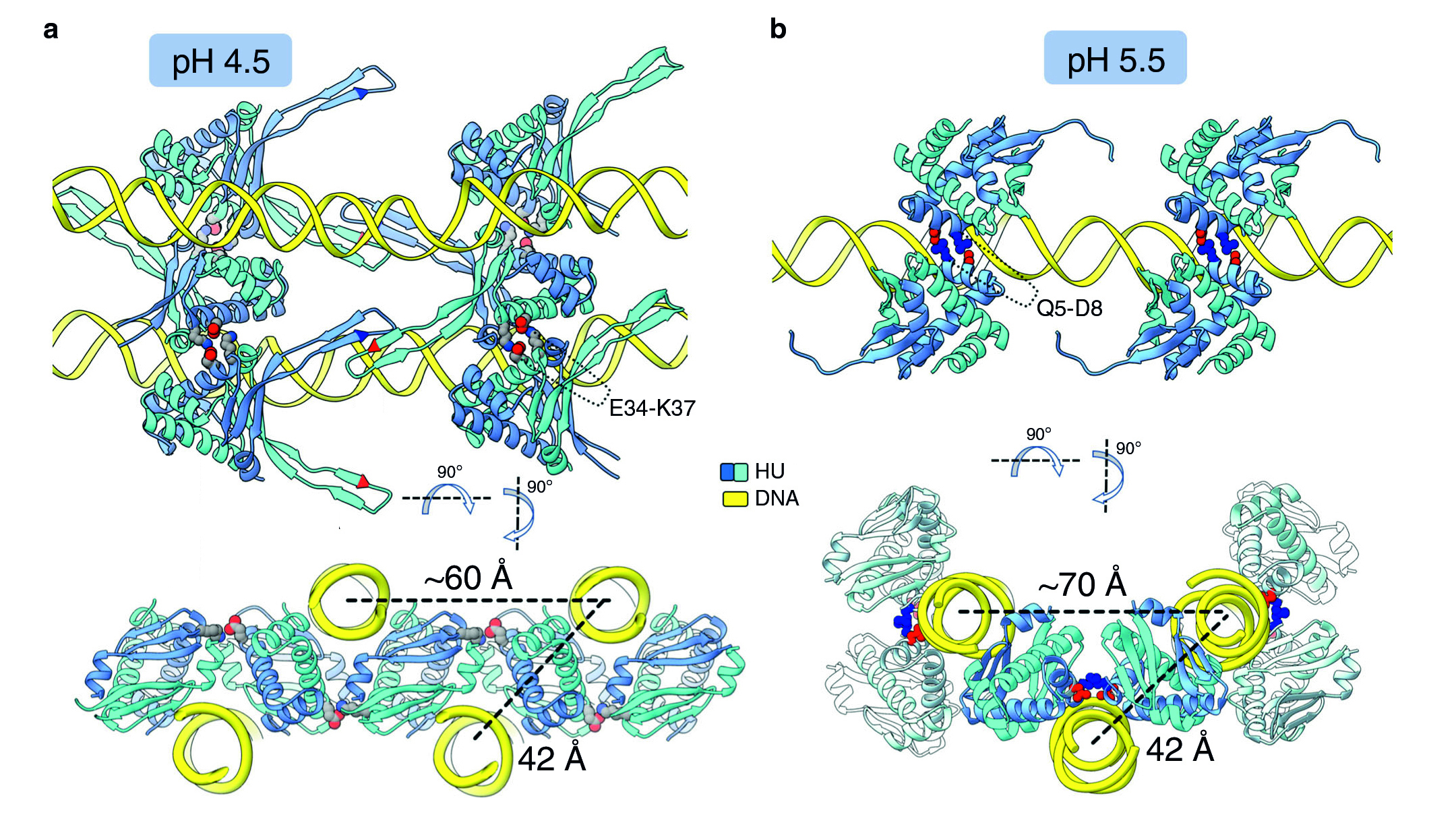
For a multiscale perspective on DNA packing by HU under various conditions (different pH levels and salt concentrations, different growth phases, and wild type vs mutated), the researchers used three x-ray visualization techniques at the ALS. At the microscale, soft x-ray tomography at Beamline 2.1 enabled characterization of the higher-order organization of the E. coli nucleoid (the bacterial analogue of a cell nucleus). At the mesoscale, small-angle x-ray scattering (SAXS) at Beamline 12.3.1 made it possible to determine the overall shapes of HU–DNA complexes in solution. At the nanoscale, protein crystallography at Beamline 8.3.1 allowed identification of the molecular-level mechanisms that mediate DNA bundling.
The combination of SAXS and crystallography was crucial to gaining a detailed understanding of the HU response to environmental changes. Ready access to all three beamlines, as well as to experts onsite, greatly facilitated the work. Finally, the researchers also performed gene expression measurements, to correlate expression levels with the tomographic and scattering data.
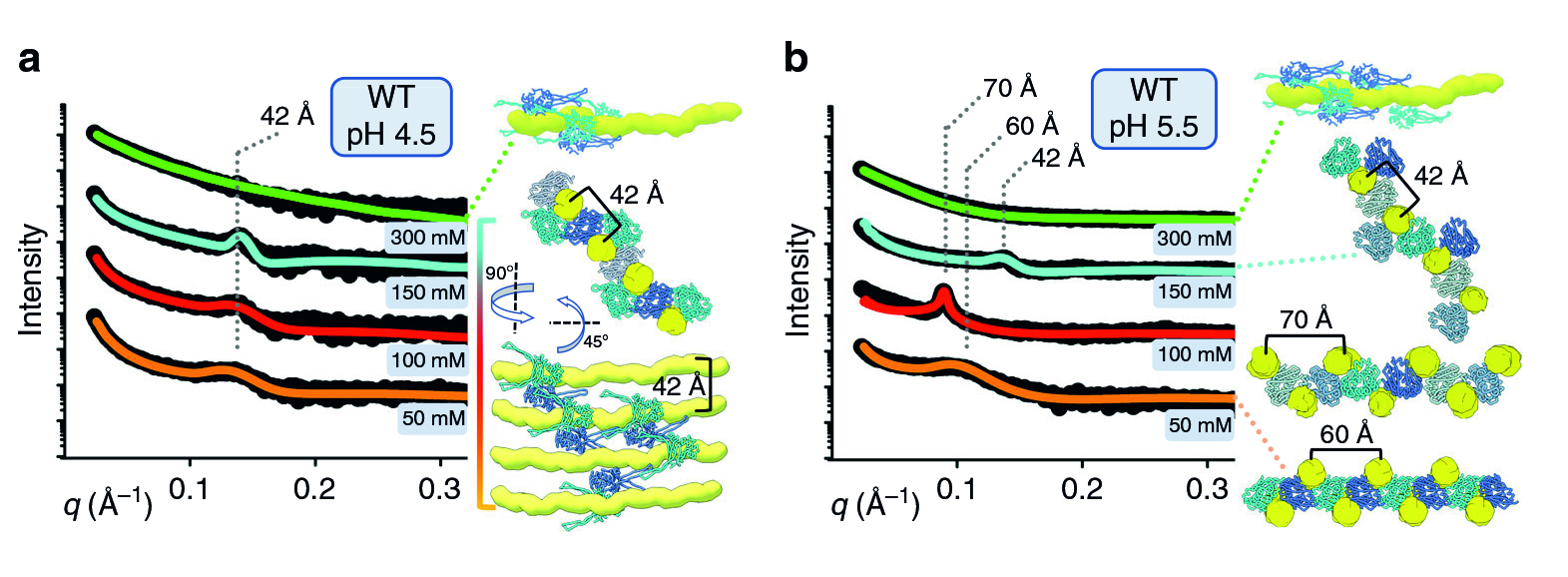
Surviving the acid test
The tomography results revealed that, under acidic conditions, the chromatin is less condensed, with more diffuse borders. This “remodeling” of the chromatin was correlated with an increase in gene expression, consistent with the need to respond quickly to survive the stress of an acidic environment. The SAXS and crystallography data showed that HU multimerization is dependent on pH and salt concentrations. These effects mechanistically rely on HU’s promiscuity in forming multiple electrostatically driven multimerization interfaces. The next step will be to figure out how to control DNA packing in order to change bacterial behavior, with the ultimate goal of developing new approaches to fighting bacterial infections.
Contact: Michal Hammel
Researchers: S.G. Remesh (National Cancer Institute and Berkeley Lab); S.C. Verma and S. Adhya (National Cancer Institute); J.-H. Chen, A.A. Ekman, and C.A. Larabell (Berkeley Lab and Univ. of California, San Francisco); and M. Hammel (Berkeley Lab).
Funding: U.S. Department of Energy (DOE), Office of Science, Biological and Environmental Research program; National Institutes of Health; University of California Office of the President; Plexxicon Inc.; and International Union of Crystallography. Operation of the ALS is supported by the DOE Office of Science, Basic Energy Sciences program.
Publication: S.G. Remesh, S.C. Verma, J.-H. Chen, A.A. Ekman, C.A. Larabell, S. Adhya, and M. Hammel, “Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling,” Nat. Commun. 11, 2905 (2020), doi:10.1038/s41467-020-16724-5.
Adapted from the Berkeley Lab news release, “Study Gains New Insight Into Bacterial DNA Packing.”
ALS SCIENCE HIGHLIGHT #426