SCIENTIFIC ACHIEVEMENT
Using the computer game, “Foldit,” nonexpert citizen scientists designed new proteins whose structures, verified at the Advanced Light Source (ALS), were equivalent in quality to and more structurally diverse than computer-generated designs.
SIGNIFICANCE AND IMPACT
The work shows the potential of using crowd-based creativity in the design of new proteins for fighting illness and disease.
A vast, unexplored protein space
Proteins constitute the biomachinery—the cellular gears and levers—that make our bodies work. When this machinery is running smoothly, nutrients get absorbed, cells regenerate, and so on. When the machinery breaks down, the tools needed to fix the problem (i.e. drug molecules) are often proteins themselves.
Until recently, the pool of proteins available for such therapeutic purposes was limited to those found in nature. But natural proteins represent a small subset of all the possible ways to link 20 amino acids—the basic building blocks of all proteins—into chains hundreds, even thousands, of units long. On top of this, there are countless ways in which any given protein chain can fold—a key aspect of functionality.
In the last 20 years, “de novo” protein design (from scratch as opposed to starting with a known protein) has taken off, promising cheap and effective drugs with fewer side effects. But given the huge number of possibilities available, scientists are limited in their ability to fully explore this vast “protein space.”
Gamification: from Rosetta to Foldit
In 2005, researchers from the University of Washington rolled out Rosetta@home, a screen-saver program that volunteers could download to their personal computers to perform protein-folding calculations in the background, while their processors were idle. Soon, volunteers began asking for a more active role, one where they could manipulate the protein folds themselves, maybe even tweak folds that Rosetta was analyzing.
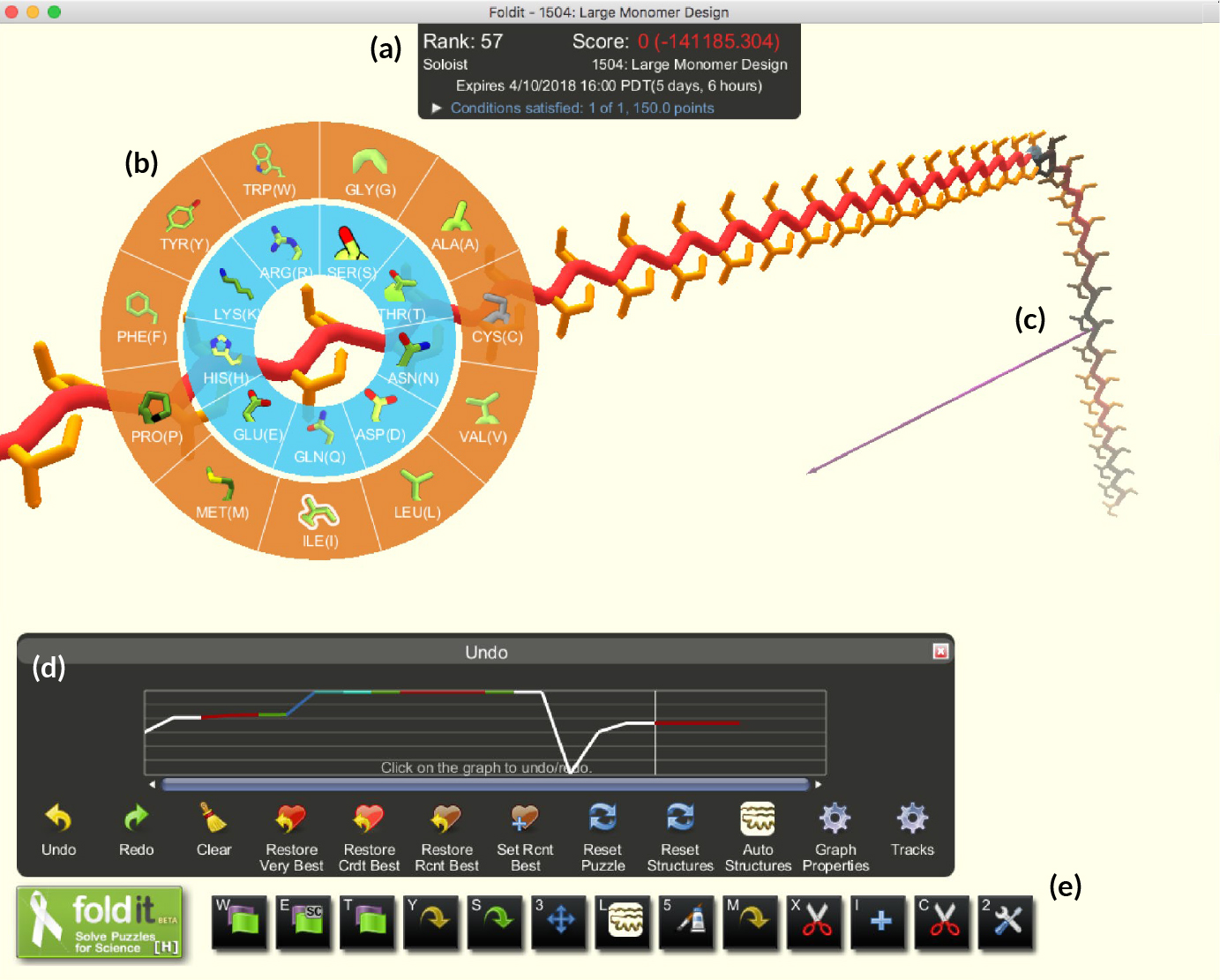
The result was the free online computer game Foldit, first introduced in 2008 and refined over time. Foldit incorporates principles of protein folding by giving higher scores for things like removing voids, covering hydrophobic strands, and clearing clashes between amino-acid side chains. In general, the lower the energy of a configuration (as calculated by Rosetta modules running behind the scenes), the higher the score. Because the game encodes the specialized knowledge of experts on molecular interactions, players are free to let their imaginations (and competitive spirit) guide their decisions.
De novo design and verification
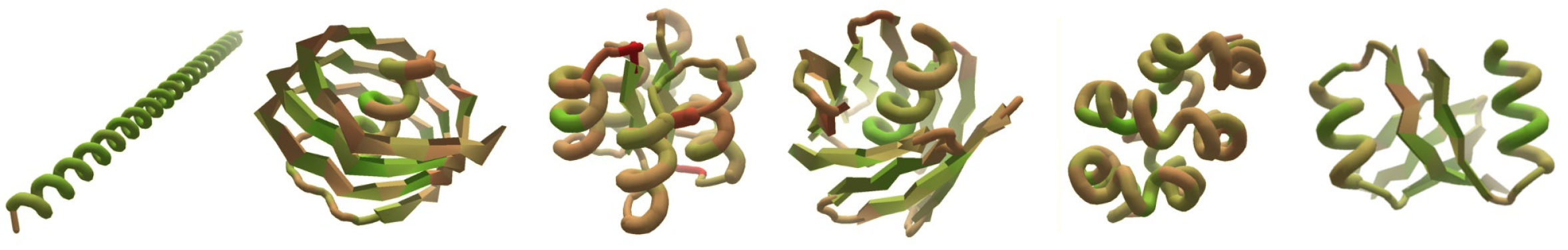
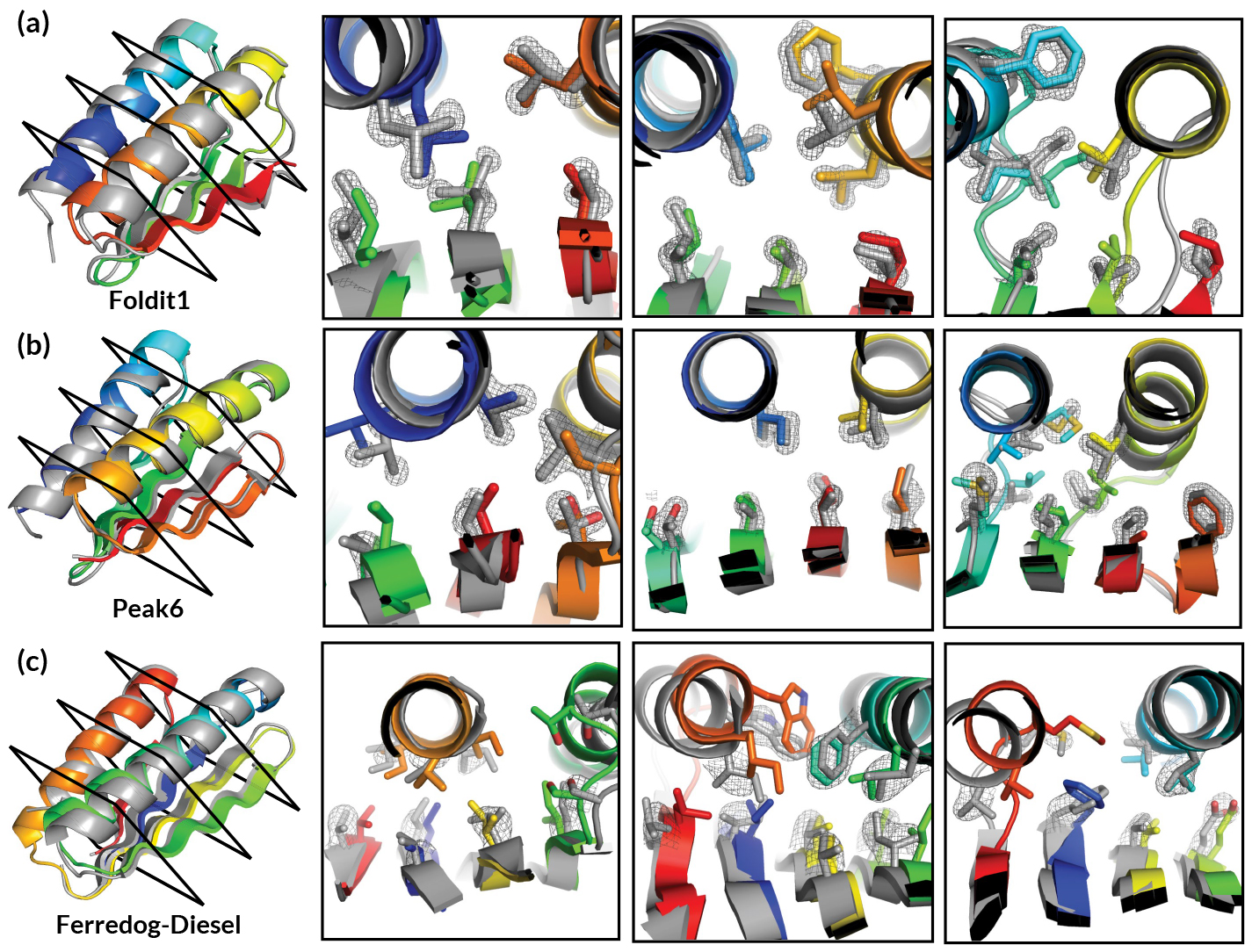
In the past, Foldit was primarily applied to the prediction or modification of known protein structures. In this work, researchers presented players with a far more expansive challenge: given a completely unfolded amino-acid chain, craft a de novo protein structure. The challenge resulted in 146 designs, 56 of which were found to adopt stable structures when produced in a lab. Four of the structures were verified experimentally—three of them using x-ray crystallography at ALS Beamlines 5.0.2, 8.2.1, and 8.2.2.
The Foldit designs—crafted by players with little or no biochemical training—turned out to be as good as those generated by experts or computers, with an equivalent success rate in the lab and similar folding stabilities. However, the researchers observed that Foldit designs are more structurally diverse: players are often willing to undergo large increases in energy to explore whole new regions in protein space.
Having established that gamers can accurately design folded proteins, the researchers next plan to present more difficult design problems, such as self-assembling protein complexes or novel proteins that bind to therapeutic targets.
Contact: Brian Koepnick
Researchers: B. Koepnick, J. Flatten, T. Husain, A. Ford, D.-A. Silva, M.J. Bick, A. Bauer, F. DiMaio, and Z. Popović (University of Washington); G. Liu (Rutgers University and Nexomics Biosciences); Y. Ishida and G.T. Montelione (Rutgers University); A. Boykov, R.D. Estep, S. Kleinfelter, T. Nørgård-Solano, L. Wei, and Foldit Players (unaffiliated); F. Khatib (University of Massachusetts Dartmouth); S. Cooper (Northeastern University); and D. Baker (University of Washington and Howard Hughes Medical Institute).
Funding: National Science Foundation, National Institutes of Health, and Howard Hughes Medical Institute. Operation of the ALS is supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences Program.
Publication: B. Koepnick, J. Flatten, T. Husain, A. Ford, D.-A. Silva, M.J. Bick, A. Bauer, G. Liu, Y. Ishida, A. Boykov, R.D. Estep, S. Kleinfelter, T. Nørgård-Solano, L. Wei, Foldit Players, G.T. Montelione, F. DiMaio, Z. Popović, F. Khatib, S. Cooper, and D. Baker, “De novo protein design by citizen scientists,” Nature 570, 390 (2019), doi:10.1038/s41586-019-1274-4.
ALS SCIENCE HIGHLIGHT #404