David Baker, a biochemistry professor at the University of Washington, head of the Institute for Protein Design, and longtime user of the Advanced Light Source (ALS), was awarded the 2024 Nobel Prize in Chemistry on October 8, alongside Demis Hassabis and John M. Jumper from DeepMind, for their groundbreaking work in protein structure prediction and design. This momentous announcement highlights significant advancements in biochemistry that have far-reaching benefits for human health and environmental sustainability.
Proteins are the molecular machines that power all living organisms, from plants to animals and humans. Baker’s work has addressed one of the most significant challenges in science: deciphering protein structure and function by designing the first de novo protein, one never seen in nature.
His team used their knowledge of how amino acid chains fold into three-dimensional structures and designed proteins that have not yet evolved, opening new avenues in medical research for diseases like Alzheimer’s and environmental solutions such as plastic bioremediation. Protein design is paving the way for pandemic deterrence, cancer therapeutics, breaking down pollutants, and carbon sequestration.
Breakthrough at the beamline
For over two decades, Baker has used the ALS synchrotron user facility at Lawrence Berkeley National Laboratory (Berkeley Lab), among other Department of Energy (DOE) Office of Science user facilities around the US and around the world, to crack the code on protein design. The ALS’s advanced high-throughput small-angle x-ray scattering (SAXS) capabilities and crystallography program supported key advancements in both experimental and computational protein science that led to the breakthroughs for Baker, Hassabis, and Jumper.
“Baker’s team has developed proteins with exceptional catalytic properties, self-assembling nanomaterials, and macromolecular switches and sensors. To verify and further optimize these designs, they rely heavily on SAXS and protein crystallography capabilities at the ALS,” said Greg Hura, science deputy of the Molecular Biophysics and Integrated Bioimaging (MBIB) Division at Berkeley Lab.
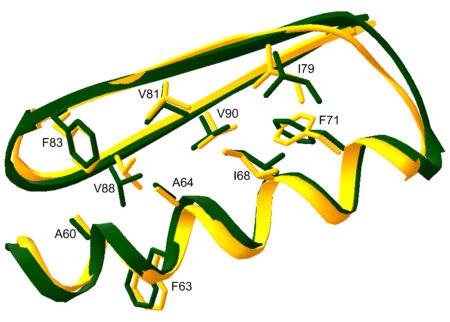
The collaboration between Baker and the ALS began in the early 2000s when the Howard Hughes Medical Institute (HHMI) provided funding for the construction and operation of macromolecular crystallography beamlines at the ALS to support HHMI principal investigators and group leaders in x-ray diffraction. In 2003, Baker collaborated with the University of North Carolina, Chapel Hill, and the Fred Hutchinson Cancer Research Center to develop a methodology for protein-structure prediction and design by creating a new type of artificial globular protein made up of 93 amino acids called Top7. This achievement, cited as a key breakthrough by the Nobel committee, was validated using the ALS HHMI Beamline 8.2.1. The data collected determined the structure of Top7 in comparison to a computationally designed model. The success of Top7 indicated two significant findings: First, it confirmed that we understand how proteins behave at an atomic level. Second, it showed that scientists can create new proteins that don’t exist in nature, which could lead to new types of medicines or machines made from proteins.
Collaborative crystallography takes shape
Over the years, Baker published 78 papers that made use of ALS beamlines, a dozen of which were highlighted by the ALS. The predicted protein structures were then verified using facilities at the ALS, where Baker has been a regular user of the Biosciences-run Berkeley Center for Structural Biology (BCSB) Beamlines and Structurally Integrated BiologY for Life Sciences (SIBYLS) Beamline for more than 20 years, contributing to nearly 166 structures deposited into the Protein Data Bank.

Banu Sankaran, an MBIB research scientist and a crystallographer at the ALS, began working with Baker in 2012 as part of the established Collaborative Crystallography Program to assist his group with x-ray crystallography. Sankaran noted, “The engineering cycle for proteins requires regular feedback between design and experiment—it was my privilege to carefully collect data, process it, and address data pathologies with the Baker group for over a decade, providing them with the data needed to move their research along.” In collaboration with David Baker, Sankaran collects and processes diffraction data on the HHMI-funded Sector 8 beamlines.
Propelling protein engineering
The ALS contributes to the “test and improve” phase of the protein engineering process by coupling bright synchrotron light with robotics at the SIBYLS SAXS beamline and BCSB/HHMI Beamlines 8.2.1 and 8.2.2. DOE’s Biological and Environmental Research program funded SIBYLS through the Integrated Diffraction Analysis Technologies (IDAT) program to build a high-throughput SAXS capability, available exclusively at the ALS facility, to support protein engineering. Over the years, the Baker lab has leveraged this capability to collect nearly 3,000 samples for rapid feedback on design. Select designs were further verified by higher-resolution crystallography and pinpoint accuracy on potential improvements. Since 2015, the IDAT team’s data has contributed to 29 of Baker’s papers, with several team members co-authoring.
Demis Hassabis and John M. Jumper developed the deep-learning approach for predicting protein structure from amino acid sequences, AlphaFold. Since the SAXS technique plays a significant role in protein engineering, it was also trialed as a rapid test of structure prediction accuracy. Susan Tsutakawa, MBIB Structural Biology department head, and Greg Hura, both members of the SIBYLS/IDAT team, were involved in assessing structure prediction efforts for the Critical Assessment of Protein Structure Prediction (CASP) experiment, where researchers are regularly challenged to predict protein structures, given only the amino acid sequences. These efforts helped recognize AlphaFold’s breakthrough during CASP13 and 14, laying the groundwork for solving the protein structure prediction problem.
Harnessing beamlines, supercomputing, and science networks
The collaboration also spans various Berkeley Lab user facilities. The Collaborative Crystallography program relies on supercomputing facilities like the National Energy Research Scientific Computing Center (NERSC), a high-performance computing research facility, for data dissemination. ESnet, the Department of Energy’s dedicated science network stewarded by Berkeley Lab, also plays an important role in linking synchrotron sites with NERSC and providing the most efficient way to transport terabytes of data between user facilities and home laboratories.
“Baker’s group at the Institute for Protein Design was one of the first to utilize this service model, and now we offer these services to 20 user groups,” stated Sankaran. As scientists harness the intricacies of protein structure and function, the benefits to society could be monumental, addressing critical challenges in health and climate change.
Just four years after congratulating Jennifer Douda on her Nobel Prize in Chemistry for her CRISPR-Cas9 crystallographic work, the ALS is proud to now commend Baker. Both awards highlight the ALS’s role in supporting a highly successful hard x-ray program at a facility optimized for soft x-rays, showcasing the scientific breakthroughs made possible through cutting-edge technology and collaborative research. These efforts are advancing the future of protein design and global science.