SCIENTIFIC ACHIEVEMENT
Using a computer-based approach, researchers designed porous protein crystals that were revealed to be stable, tunable, and atomically accurate using x-ray scattering and diffraction at the Advanced Light Source (ALS).
SIGNIFICANCE AND IMPACT
The work provides a powerful new platform for biological materials engineering and opens up wide applications in biotechnology and medicine.

Precise and predictable crystal design
Protein crystallization has always been mysterious—requiring a lot of work and luck to coax large, irregular molecules to form orderly crystalline arrays. Typically, scientists have wanted to crystallize protein molecules to learn where the atoms are, using x-ray diffraction, to understand or manipulate protein function. Now, with computational protein design, scientists are exploring ways to use proteins to create completely new materials with precisely tuned characteristics such as lattice dimensions and pore sizes.

In this work, an international team led by researchers from the Institute for Protein Design at the University of Washington (UW) developed a computer-aided approach for designing three-dimensional protein crystals with prespecified lattice architectures and atomic accuracy. The ability to genetically encode and express precise building blocks for synthesized biomaterials sets the stage for the development of advanced optical tools, new technologies for chemical separation, and a range of other applications in biotechnology and medicine.
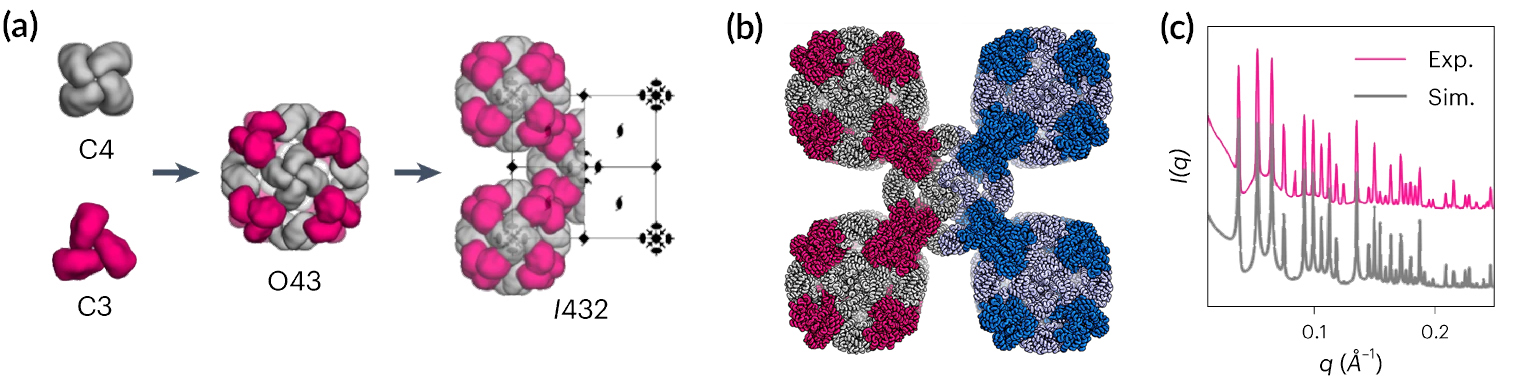
Hierarchical approach with protein modules
Protein crystallization remains poorly understood and highly empirical, with largely unpredictable crystal contacts, lattice packing arrangements, and symmetry properties. One challenge is to precisely engineer interactions between side chains across protein interfaces. Each interface must have high geometric precision, as small deviations can add up over multiple unit cells. Previously, the UW team used computational methods to design protein building blocks and assemblies that were structurally characterized at the ALS, including repeat proteins, hydrogen-bonded helical bundles, cyclic oligomers, and protein nanocages.
In this work, the team followed a hierarchical approach, designing protein modules (oligomers) made up of monomers with programmed sequences, expressed through bacteria. The oligomers then self-assemble into cages and, ultimately, three-dimensional crystals hundreds of microns in size. The team also encapsulated gold nanoparticles within the crystal pores, creating a nanoparticle superlattice with dynamic optical properties upon drying and rehydration.
Screening and verification at the ALS
High-throughput small-angle x-ray scattering (SAXS) at ALS Beamline 12.3.1 (SIBYLS) was used to screen and fine-tune proteins to fold into desired structures. SAXS characterization continued through the assembly process to the final macroscale crystals. Protein crystallography at ALS Beamlines 8.2.1 and 8.2.2 and at the Advanced Photon Source (APS) verified that the experimental crystals matched the designs with high precision, both in overall architecture and specific protein–protein interactions.
A custom SAXS setup, developed for the crystals with gold nanoparticles, demonstrated that crystallinity was maintained during drying and rehydration cycles. Simultaneously varying the hydration state and the encapsulation ratio enabled tuning of the crystal’s effective refractive index (and thus the optical properties) over a broad range.
The study exemplifies the application of Integrated Diffraction Analysis Technologies (IDAT) resources (a grant to Berkeley Lab from the Biological and Environmental Research program of the Department of Energy), funded to integrate SAXS, computation, and crystallography for novel biomaterial discovery. In general, the work demonstrates that computationally designed protein crystals are a new class of biomaterials that can be prepared and purified at large scale, are stable under extreme conditions, and provide robust and tunable scaffolds for materials enabling a wide range of applications such as biocatalysis, sensing, chemical separation, and drug delivery.
Contact: Zhe Li
Researchers: Z. Li, S. Wang, U. Nattermann, A.K. Bera, A.J. Borst, M.Y. Yaman, M.J. Bick, E.C. Yang, W. Sheffler, H. Nguyen, A. Kang, R. Dalal, J.M. Lubner, Y. Hsia, H. Haddox, Q. Dowling, M. Miranda, A. Favor, N.I. Edman, W. Yang, C. Weidle, and D.S. Ginger (University of Washington); B. Lee and S. Seifert (Argonne National Laboratory); G.L. Hura and B. Sankaran (Berkeley Lab); A. Courbet and D. Baker (University of Washington and Howard Hughes Medical Institute); A. Etemadi (University of Washington and Tehran University of Medical Sciences); B. Negahdari (Tehran University of Medical Sciences), and M.B. Ross (University of Massachusetts Lowell).
Funding: Howard Hughes Medical Institute; Amgen; Novo Nordisk; Institute for Protein Design Directors Fund; Audacious Project at the Institute for Protein Design; Open Philanthropy Project Improving Protein Design Fund; Defense Advanced Research Projects Agency; Washington Research Foundation; Nordstrom Barrier Institute for Protein Design Directors Fund; Human Frontiers Science Program; National Institutes of Health; National Science Foundation; US Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES) program; DOE Office of Science, Biological and Environmental Research program, IDAT grant; and Center for the Science of Synthesis Across Scales (a DOE BES Energy Frontier Research Center). Operation of the ALS and APS is supported by DOE BES.
Publication: Z. Li, S. Wang, U. Nattermann, A.K. Bera, A.J. Borst, M.Y. Yaman, M.J. Bick, E.C. Yang, W. Sheffler, B. Lee, S. Seifert, G.L. Hura, H. Nguyen, A. Kang, R. Dalal, J.M. Lubner, Y. Hsia, H. Haddox, A. Courbet, Q. Dowling, M. Miranda, A. Favor, A. Etemadi, N.I. Edman, W. Yang, C. Weidle, B. Sankaran, B. Negahdari, M.B. Ross, D.S. Ginger, and D. Baker, “Accurate Computational Design of 3D Protein Crystals,” Nat. Mater. 22, 1556 (2023), doi: 10.1038/s41563-023-01683-1.
Adapted from the Institute for Protein Design (UW) article, “Designing protein crystals with sub-nanometer accuracy.”
ALS SCIENCE HIGHLIGHT #495